Abstract
Previously, a pathogenic ciliate was isolated from the surface ulcer of a diseased Turbot (Scophthalmus maximus L.) at an aquaculture farm in North China. After morphological and molecular biological identification based on 18rRNA, the ciliated was identified as the notorious scuticociliates (Pseudocohnilembus persalinus). In this study, the whole sequence of the mitochondrial genomic gene of P. persalinus was carried out. The sequencing results showed that the complete sequence of P. persalinus mitogenome was 38,375 bp. There were 2 rRNAs, 4 tRNAs, and 34 protein-coding genes (PCGs), respectively, located on both the heavy strand and the light stand. 15 PCGs were on the heavy strand, and 19 PCGs on the light strand. Besides, phylogenetic trees among 11 ciliates were constructed based on the sequences of 17 PCGs located in mitogenome using BI methods. The results of clustering showed that P. persalinus and Uronema marinum was the first cluster belonging to the order Scuticociliatida. Our research results will further provide primary data for evolution and classified study of scuticociliates.
Turbot (Scophthalmus maximus L.) was a fast-growing, highly cold tolerance flatfish with high economic value, and had already become main factory-farmed species in North China (Lei et al. Citation2005). However, with the expansion of the size of Turbot farming, diseases followed, among which scuticociliatosis was especially serious in recent years (Cui et al. Citation2008). Pseudocohnilembus persalinus was very notorious as a representative scuticociliate (Kim et al, Citation2004).
In this study, the whole sequence of the mitochondrial genome of P. persalinus, previously isolated from the surface ulcer of a diseased Turbot (S. maximus L.) at an aquaculture farm in North China (38.9768 N, 121.3459 E), was sequenced and assembled by Illumina MiSeq Next-generation sequencing by SC Gene Company (Guangzhou, China). The sequence of P. persalinus mitochondrial genome was submitted and deposited in the GenBank (accession number MH608212). The results showed that the complete sequence of P. persalinus mitogenome was 38,375 bp. There were 2 rRNAs, 4 tRNAs (tRNA-Glu, tRNA-Phe, tRNA-His, and tRNA-Trp), and 34 protein-coding genes (PCGs), respectively, located on both the heavy strand and the light stand. 15 PCGs were on the heavy strand, and 19 PCGs on the light strand. Besides, 14 genes (rpl14, ymf70, nad4, rps3, nad10, nad7, rps14, ymf75, atp9, nad3, cob, cox2, ymf56, and ymf67) used the start codon ATG. Three genes (cox1, rps13, and ymf64) used the start codon ATA. Eight genes (nad1_a, nad6, ymf66, rps19, nad2, ymf65, nad4l, and ymf68) used the start codon TTA. Four genes (rpl2, ymf63, yejR and nad5) used the start codon ATT. Three genes (rps12, rpl6, and rpl16) used the start codon ATA. nad1_b used the start codon CTG. nad9 used the start codon TAT. ymf65 and ymf64 used the stop codon TAG, and all other 32 PCGs used stop codon TAA.
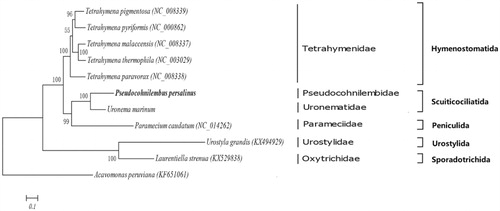
Phylogenetic trees among 11 ciliates were constructed based on the sequences of 17 PCGs (atp9, cox1, cox2, cytb, nad10, nad1_a, nad1_b, nad7, nd3, nd4, rpl14, rpl16, rpl2, rps12, rps13, rps14 and rps19) located in mitogenome using BI and ML methods. The results of clustering showed that P. persalinus and Uronema marinum was the first cluster belonging to the order Scuticociliatida (). Moreover, U. marinum was also a representative scuticocilaite isolated from Takifugu rubripes previously (Li et al. Citation2018). The other ciliates were in obvious clades: Hymenostomatida, Peniculida, Urostylida, and Sporadotrichida ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Cui LB, Zhou XY, Cheng JW, Li YS, Ma JH. 2008. Pathology of scuticociliatosis in Scophthalmus maximus L. J Fish Sci China. 15:622–629.
- Kim SM, Cho JB, Lee EH, Kwon SR, Kim SK, Nam YK, Kim KH. 2004. Pseudocohnilembus persalinus (Ciliophora: Scuticociitida) is an additional species causing scuticociliatosis in olive flounder Paralichthys olivaceus. Dis Aquat Org. 62:239.
- Lei JL, Ma AJ, Chen C, Zhuang ZM. 2005. The present status and sustainable development of turbot (Scophthalmus maximus L.) culture in China. Eng Sci. 7:30–34.
- Li RJ, Gao YQ, Hou YL, Ye SG, Wang LS, Sun JX, Li Q. 2018. Mitochondrial genome sequencing and analysis of scuticociliates (Uronema marinum) isolated from Takifugu rubripes. Mitochondrial DNA Part B. 3:736–737.