Abstract
The complete mitochondrial genome of the female-wingless bagworm moth, Eumeta variegata Snellen, 1879 (Lepidoptera: Psychidae), is 15,660 base pairs (bp) and contains a typical set of genes (13 protein-coding genes [PCGs], 2 rRNA genes, and 22 tRNA genes) and one non-coding region, with an arrangement identical to that observed in most lepidopteran genomes. Twelve PCGs contained the typical ATN start codon, whereas COI had the atypical CGA codon, which is frequently detected in the start region of the lepidopteran COI. The A + T-rich region was unusually short with only 94 bp. A recent report of the same species originating from Japan revealed a lack of trnE and trnF and a 1,118 bp long A + T-rich region. Phylogenetic analyses with concatenated sequences of the 13 PCGs and two rRNA genes using the Bayesian inference method placed E. variegata in Psychidae, as a sister to a within-familial species, Mahasena colona, with the highest nodal support (Bayesian posterior probability = 1).
The female-wingless bagworm moth, Eumeta variegata Snellen, 1879 (Lepidoptera: Psychidae), is distributed in Australia and Asia, including in Korea (Turner Citation1947; Robinson et al. Citation1994; Sobczyk Citation2011). In Korea, the species was named as E. japonica Heylaerts, 1884, but it has recently been synonymized as E. variegata (Roh et al. Citation2016). The larvae of this species make case for larval and pupal stages (Zhang Citation1997; Gries et al. Citation2006). Although the adult males have wings, female wings are reduced or missing, and thus the females and larvae have a similar appearance (Niitsu et al. Citation2008).
An adult male E. variegata was collected from Gageodo, Jeollanamdo Province (34°04′38″N, 125°06′10″E), South Korea in 2013. This voucher specimen was deposited at the Chonnam National University, Gwangju, Korea, under accession no. CNU5692. Using DNA extracted from the hind legs, three long overlapping fragments (LFs; COI-ND4, ND5-lrRNA, and lrRNA-COI) were amplified using previously described primers (Kim et al. Citation2012). These three LFs were used as templates to amplify 26 short fragments (Kim et al. Citation2012).
Phylogenetic analysis using the concatenated nucleotide sequences of 13 protein-coding genes (PCGs) and two rRNA genes was performed using a Bayesian inference (BI) method implemented in CIPRES Portal v. 3.1 (Miller et al. Citation2010). An optimal partitioning scheme (six partitions) and substitution model (GTR + Gamma + I) were determined using PartitionFinder 2 and the Greedy algorithm (Lanfear et al. Citation2012, Citation2014, Citation2016).
The complete 15,660 base pair (bp) mitochondrial genome (mitogenome) of E. variegata was composed of typical gene sets (two rRNAs, 22 tRNAs, and 13 PCGs) and a major non-coding A + T-rich region (GenBank accession no. MH574939). The length of the E. variegata A + T-rich region was the shortest at 94 bp among sequenced Tineoidea (728–1610 bp; data not shown). The gene arrangement of E. variegata is identical to that of the ditrysian Lepidoptera with trnM-trnI-trnQ between the A + T-rich region and ND2 junction (Kim et al. Citation2010). Twelve PCGs contained the typical ATN start codon, whereas COI showed an atypical CGA codon frequently found in the start region of the lepidopteran COI (Kim et al. Citation2010). Recently, the complete mitogenome of the same species originating from Japan was reported (Arakawa et al. Citation2018). The overall sequence identity between the E. variegata described in this study and that from Arakawa et al. (Citation2018) for PCGs and rRNAs was high, ranging from 97.81% (srRNA) to 99.39% (CytB) after removing non-overlapping sequences at the beginning and end of each gene. However, the two sequences differed in gene content; the species described by Arakawa et al. (Citation2018) contained only 20 tRNAs, and was missing trnE and trnF. Further, the A + T-rich region of our E. variegata mitogenome sequence was 94 bp, whereas that of the species described by Arakawa et al. (Citation2018) was 1118 bp, including 121 bp eight repeat sequences. Our 94 bp A + T-rich region sequence was well-aligned at the beginning and end regions with the sequence reported by Arakawa et al. (Citation2018).
Figure 1. Phylogenetic tree of Ditrysia (Tineoidea, Gracillarioidea, Yponomeutoidea), including E. variegata. The tree was constructed using nucleotide sequences of 13 protein-coding genes and two rRNAs with the Bayesian inference (BI) method. The numbers at each node specify Bayesian posterior probabilities. Scale bar indicates the number of substitutions per site. Astrotischeia sp. (Nepticuloidea; Tischeriidae) and Stigmella roborella (Nepticuloidea; Nepticulidae) were used as outgroups. GenBank accession numbers are as follows: Mahasena oolona, KY856825 (Li et al. Citation2017); Tineola bisselliella, KJ508045 (Timmermans et al. Citation2014); Phyllonorycter platani, KJ508044 (Timmermans et al. Citation2014); P. froelichiella, KJ508048 (Timmermans et al. Citation2014); Cameraria ohridella, KJ508042 (Timmermans et al. Citation2014); Prays oleae, KM874804 (van Asch et al. Citation2014); E. variegata, MH574939 (This study); Leucoptera malifoliella, JN790955 (Wu et al. Citation2012); Plutella xylostella, JF911819 (Wei et al. Citation2013); P. xylostella, KM023645 (Dai et al. Citation2016); Astrotischeria sp., KJ508056 (Timmermans et al. Citation2014); and Stigmella roborella, KJ508054 (Timmermans et al. Citation2014).
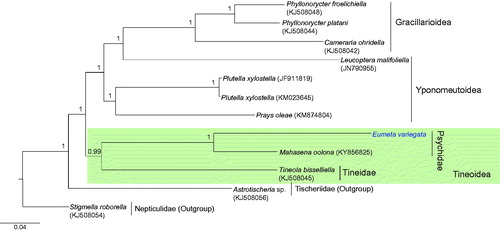
Phylogenetic analysis revealed a sister relationship between E. variegata and the within-familial species Mahasena colona (Li et al. Citation2017) with the highest nodal support (Bayesian posterior probability = 1). Currently, mitogenome sequences are available for only three species in two families in Tineoidea, including E. variegata. Thus, additional mitogenome sequences from a diverse taxonomic group are required to infer the relationships among families in Tineoidea.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arakawa K, Kono N, Ohtoshi R, Hiroyuki N, Tomita M. 2018. The complete mitochondrial genome of Eumeta variegata (Lepidoptera: Psychidae). Mitochondrial DNA B Resour. 3:812–813.
- Dai LS, Zhu BJ, Qian C, Zhang CF, Li J, Wang L, Wei GQ, Liu CL. 2016. The complete mitochondrial genome of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Mitochondrial DNA Part A. 27:512–1513.
- Gries R, Khaskin G, Tan ZX, Zhao BG, King GGS. 2006. (1S)-1-ethyl-2-methylpropyl dimethyl pentadecanoate: major sex pheromone component of Paulownia bagworm, Clania variegata. J Chem Ecol. 32:1673–1685.
- Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, Han YS, Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 15:162–173.
- Kim MJ, Wan X, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Li PW, Chen SC, Xu YM, Wang XQ, Hu X, Peng P. 2017. The complete mitochondrial genome of a tea bagworm, Mahasena colona (Lepidoptera: Psychidae). Mitochondrial DNA Part B. 2:381–382.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), IEEE, Nov 14; 1–8; New Orleans
- Niitsu S, Lobbia S, Izumi S, Fujiwara H. 2008. Female-specific wing degeneration is triggered by ecdysteroid in cultures of wing discs from the bagworm moth, Eumeta variegata (Insecta: Lepidoptera, Psychidae). Cell Tissue Res. 333:169–173.
- Robinson GS, Tuck KR, Shaffer M. 1994. Smaller moths of Southeast Asia. London: Natural History Museum
- Roh SJ, Banasiak G, Byun B-K. 2016. A new and an unrecorded species of the family Psychidae (Lepidoptera) from Korea, with an annotated catalogue. J Nat Hist. 50:669–680.
- Sobczyk T. 2011. Psychidae (Lepidoptera): world catalogue of insects. Stenstrup: Apollo Books.
- Timmermans MJ, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178.
- Turner AJ. 1947. A review of the phylogeny and classification of the Lepidoptera. Proc Linn Soc NSW. 71:303–338.
- van Asch B, Blibech I, Pereira-Castro I, Rei FT, da Costa LT. 2014. The mitochondrial genome of Prays oleae (Insecta: Lepidoptera: Praydidae). Mitochondria DNA. 27:1–2.
- Wei SJ, Shi BC, Gong YJ, Li Q, Chen XX. 2013. Characterization of the mitochondrial genome of the diamondback moth Plutella xylostella (Lepidoptera: Plutellidae) and phylogenetic analysis of advanced moths and butterflies. DNA Cell Biol. 32:173–187.
- Wu YP, Zhao JL, Su TJ, Li J, Yu F, Chesters D, Fan RJ, Chen MC, Wu CS, Zhu CD. 2012. The complete mitochondrial genome of Leucoptera malifoliella Costa (Lepidoptera: Lyonetiidae). DNA Cell Biol. 31:1508–1522.
- Zhang ZZ. 1997. Forest entomology. Beijing: China Forestry Publishing House.
