Abstract
Hydnocarpus hainanensis Merr. is an evergreen tree with a height of 6–12 m and a diameter at breast height of 50 cm. It is distributed in Guangxi, Guizhou, Hainan, South of Yunnan Province of China. Here, we report and characterize the complete plastid genome sequence of H. hainanensis in an effort to provide genomic resources useful for promoting its conservation and systematics research. The plastome of H. hainanensis is found to possess a total length 163,330 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 26,870 bp, a large single copy (LSC) region of 91,510 bp and a small single copy (SSC) region of 18,080 bp. The plastome contains 111 genes, consisting of 78 unique protein-coding genes (seven of which are duplicated in the IR: rps12, rps7, ndhB, ycf2, rpl23, rpl2, and rps19), 29 unique tRNA genes (seven of which are duplicated in the IR, i.e. trnNGUU, trnRACG, trnAUGC, trnlGAU, trnVGAC, trnLCAA, and trnlCAU) and four unique rRNA genes (5S rRNA, 4.55S rRNA, 23S rRNA, and 16S rRNA). The overall A/T content in the plastome of H. hainanensis is 63.70%. The phylogenetic analysis indicated that H. hainanensis is close to Salix rorida within Malpighiales. The complete plastome sequence of H. hainanensis will provide a useful resource for the conservation genetics of the one species as well as for the phylogenetic studies of Achariaceae.
Introduction
Hydnocarpus hainanensis is an evergreen tree with a height of 6–12 m and a diameter at breast height of 50 cm. It is distributed throughout the evergreen broad-leaved forests having altitudes that range from 300 to 1800 m in Guangxi, Guizhou, Hainan, South of Yunnan Province in China (Yang and Sue Citation2007). It has been ranked as a VU (vulnerable) species in China (Ministry of Environmental Protection of the People’s Republic of China and Chinese Academy of Sciences Citation2013). In China, this species is under threat because of its habitat loss and harvesting of the durable and decay-resistant timber and the fruit for the treatment of skin conditions. Besides, its natural regeneration is poor. Consequently, its genetic and genomic information are urgently needed in order to promote its conservation and economic use of H. hainanensis. Here, we report and characterize the complete plastome of H. hainanensis (GenBank accession number: MH708163, this study). This is the first report of a complete plastome for the H. hainanensis.
In this study, H. hainanensis is sampled from Diaoluo Mountain (18.67°N, 109.88°E), which is a National Nature Reserve of Hainan, China. The voucher specimens (Wang et al. B50) are deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
The modified CTAB method of Doyle and Doyle (Citation1987) is used to extract total genomic DNA from leaves quickly frozen with dry ice. One microgram of genomic DNA is used for Illumina library preparation, using version 3 chemistry. Paired-end, 150 bp reads are sequenced using an Illumina HiSeq 2500 platform at the Guangzhou Novel-seq Biotechnology Co., Ltd. (Guangzhou, China). Reads are trimmed and those with >10% Ns or with >10% low-quality (Q ≤ 5) bases are filtered out using NGSQC-Toolkit v2.3.3 (Patel and Jain Citation2012). Cleaned reads are assembled against the plastome of Idseia polycarpa (NC_032060.1) (Yang et al. Citation2016) using MITObim v1.8 (Hahn et al. Citation2013). The plastome is annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Idseia polycarpa (NC_032060.1). The annotation is corrected with DOGMA (Wyman et al. Citation2004).
The plastome of H. hainanensis is found to possess a total length 163,330 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 26,870 bp, a large single copy (LSC) region of 91,510 bp and a small single copy (SSC) region of 18,080 bp. The plastome contains 111 genes, consisting of 78 unique protein-coding genes (seven of which are duplicated in the IR, i.e. rps12, rps7, ndhB, ycf2, rpl23, rpl2, and rps19), 29 unique tRNA genes (seven of which are duplicated in the IR, i.e. trnN-GUU, trnR-ACG, trnA-UGC, trnl-GAU, trnV-GAC, trnL-CAA, and trnl-CAU) and four unique rRNA genes (5SrRNA, 4.55SrRNA, 23S rRNA, and 16SrRNA). Among these gene, one pseudogene (ndhF, translation from 120,829 to 118,625), 13 genes (trnV-UAC, trnL-UAA, trnI-GAU, trnK-UUU, trnA-UGC, atpF, petB, petD, rpoC1, rpl16, rpl2, ndhB, and ndhA) possessed a single intron and three genes (ycf3, clpP, and rps12) had two introns. The gene rps12 is found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of H. hainanensis is 63.70%, in which the corresponding value of the LSC, SSC, and IR region is 66.10%, 69.50%, and 57.50%, respectively.
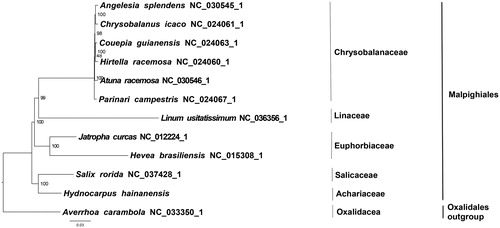
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 10 published complete plastome of Malpighiales, using Averrhoa carambola (Oxalidales, Oxalidaceae) as outgroup. The phylogenetic analysis indicated that H. hainanensis is closer to Salix rorida than other taxa within Malpighiales (). Most nodes in the plastome ML trees are strongly supported. The complete plastome sequence of H. hainanensis will provide a useful resource for the conservation genetics of the species as well as for the phylogenetic studies of Achariaceae.
Figure 1. The best ML phylogeny recovered from 12 complete plastome sequences by RAxML. Accession numbers: Hydnocarpus hainanensis (MH708163, this study), Linum usitatissimum NC_036356.1, Salix rorida NC_037428.1, Chrysobalanus icaco NC_024061.1, Parinari campestris NC_024067.1, Couepia guianensis NC_024063.1, Hirtella racemosa NC_024060.1, Hevea brasiliensis NC_015308.1, Jatropha curcas NC_012224.1, Atuna racemosa NC_030546.1, Angelesia splendens NC_030545.1; outgroup: Averrhoa carambola NC_033350.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Lee JH, Choi IS, Choi BH, Yang S, Choi G. 2016. The complete plastid genome of Piper kadsura (Piperaceae), an East Asian woody vine. Mitochondrial DNA A DNA Mapp Seq Anal. 27:3555.
- Ministry of Environmental Protection of the People’s Republic of China. 2013. The evaluation of China biodiversity red list-higher plants; [accessed 2018 Jul 17]. http://www.zhb.gov.cn/gkml/hbb/bgg/201309/W020130917614244055331.pdf.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang W, Wang W, Zhang L, Chen Z, Guo X, Ma T. 2016. Characterization of the complete chloroplast genome of Idesiapolycarpa. Conserv Genet Resour. 8:271–273.
- Yang Q, Sue Z. 2007. Flora of China: Flacourtiaceae. Beijing: Science Press and the Missouri Botanical Garden Press.
