Abstract
In this study, the complete mitochondrial genome (mitogenome) of the assassin bug Inara alboguttata, is determined using next-generation sequencing. The mitogenome is a typical circular DNA molecule of 15,436 bp long with a high AT bias (72.3%), containing 13 protein-coding genes, two rRNA genes, 22 tRNA genes and a control region. Protein-coding genes all initiate with ATN codons and most of them terminate with TAA or TAG codons, whereas COI, ND3, and ND5 use a single T residue. The lrRNA and srRNA genes are 1247 bp and 766 bp in length, respectively. All tRNA genes have the cloverleaf secondary structure except for the tRNASer(AGN). The control region is 860 bp long with an A + T content of 68.2%. Phylogenetic result supports the sister relationship between I. alboguttata and Acanthaspis ruficeps.
Inara Stål is a small genus within the subfamily Reduviinae (Hemiptera: Reduviidae) including six known species, and all of them are distributed in the Oriental regions (Maldonado-Capriles Citation1990). In this study, we characterized the complete mitochondrial genome (mitogenome) sequence of Inara alboguttata, the first representative of the genus Inara and performed an analysis of the phylogenetic relationships among Reduviidae with the available mitogenomic sequences.
Total genomic DNA was extracted from the thorax of adult collected from Nonggang National Nature Reserve in Guangxi province, China. Voucher specimen (No. VCim-00107) was deposited at the Entomological Museum of China Agricultural University (CAU). The complete mitogenome was obtained by next-generation sequencing method with Illumina Hiseq 2500 and the sequence was deposited in GenBank under the accession number KY069959.
The whole mitogenome is a circular DNA molecule of 15,436 bp long, including 37 genes (13 protein-coding genes, 22 tRNA genes and two rRNA genes) and a control region. The gene arrangement is largely the same as the putative ancestral arrangement of insects and most assassin bugs (Li et al. Citation2011, Citation2015; Gao et al. Citation2013; Zhao et al. Citation2015; Zhang et al. Citation2016). The overall nucleotide composition of the whole mitogenome is significantly biased toward A + T (72.3%) with positive AT-skew (0.14) and negative GC-skew (−0.16). All protein-coding genes initiate with ATN as the start codon (three with ATT, five with ATA and five with ATG). Conventional stop codons (TAA or TAG) have been assigned to majority of the protein-coding genes. COI, ND3, and ND5, however, terminate with a single T residue.
This mitogenome has the complete set of 22 tRNA genes, ranging from 61 to 70 bp. All of the tRNA genes have a typically cloverleaf structure except for the dihydrouridine (DHU) arm of tRNASer(AGN) forms a simple loop. The lrRNA is 1247 bp long with an A + T content of 74.5%, and the srRNA is 766 bp long with an A + T content of 72.2%. The control region located between srRNA and tRNAIle is 860 bp long with an A + T content of 68.2%. Tandem repeats of two 27-nt units are found in the control region.
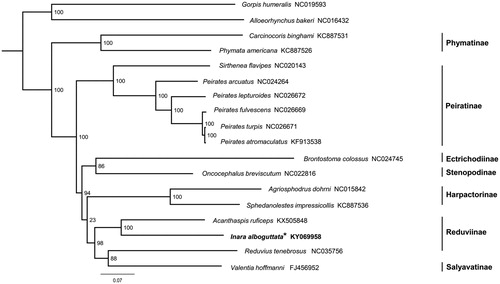
Maximum-likelihood (ML) tree was constructed based on sequences of 13 protein-coding genes and two rRNA genes from 16 Reduviidae species by using RAxML-HPC2 (Stamatakis Citation2014) under the GTR + I + G model (). The sister relationship between I. alboguttata and Acanthaspis ruficeps is highly supported. However, Reduvius tenebrosus is the sister group to Valentia hoffmanni (Salyavatinae), therefore, the monophyly of Reduviinae is not recovered, which is similar to the results of previous studies (Weirauch and Munro Citation2009; Hwang and Weirauch Citation2012; Jiang et al. Citation2016).
Figure 1. Phylogenetic tree inferred from ML analysis of the nucleotide of the 13 protein-coding genes and two rRNA genes (12,697 bp). The nodal values indicate the bootstrap percentages obtained with 1000 replicates. GenBank accession numbers for the sequences are indicated next to species name. Mitogenome newly sequenced in the present study is highlighted by the asterisk.
Disclosure statement
All authors have read and approved the final manuscript. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Gao JY, Li H, Truong XL, Dai X, Chang J, Cai WZ. 2013. Complete nucleotide sequence and organization of the mitochondrial genome of Sirthenea flavipes (Hemiptera: Reduviidae: Peiratinae) and comparison with other assassin bugs. Zootaxa. 3669:1–16.
- Hwang WS, Weirauch C. 2012. Evolutionary history of assassin bugs (Insecta: Hemiptera: Reduviidae): insights from divergence dating and ancestral state reconstruction. PLoS One. 7:e45523.
- Jiang P, Li H, Song F, Cai Y, Wang JY, Liu JP, Cai WZ. 2016. Duplication and remolding of tRNA genes in the mitochondrial genome of Reduvius tenebrosus (Hemiptera: Reduviidae). IJMS. 17:951.
- Li H, Gao JY, Cai WZ. 2015. Complete mitochondrial genome of the assassin bug Oncocephalus breviscutum (Hemiptera: Reduviidae). Mitochondrial DNA. 26:674–675.
- Li H, Gao JY, Liu HY, Liu H, Liang AP, Zhou XG, Cai WZ. 2011. The architecture and complete sequence of mitochondrial genome of an assassin bug Agriosphodrus dohrni (Hemiptera: Reduviidae). Int J Biol Sci. 7:792–804.
- Maldonado-Capriles J. 1990. Systematic catalogue of the Reduviidae of the world (Insecta: Heteroptera). A special edition of Caribbean Journal of Science. Puerto Rico. 694:407–408.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Weirauch C, Munro JB. 2009. Molecular phylogeny of the assassin bugs (Hemiptera: Reduviidae), based on mitochondrial and nuclear ribosomal genes. Mol. Phylogenet. Evol. 53:287–299.
- Zhang LJ, Song F, Li H, Cai WZ. 2016. First complete mitochondrial genome sequence from the tribelocephaline assassin bugs (Hemiptera: Reduviidae). Mitochondrial DNA A. 27:4203–4204.
- Zhao GY, Li H, Zhao P, Cai WZ. 2015. Comparative mitogenomics of the assassin bug genus Peirates (Hemiptera: Reduviidae: Peiratinae) reveal conserved mitochondrial genome organization of P. atromaculatus, P. fulvescens and P. turpis. PLoS One. 10:e0117862.
