Abstract
In this study, we first characterized the complete mitogenome of Pygathrix nigripes, and analysed its phylogenetic status. The circular mitogenome was 16,534 bp in length, and contained 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes and one non-coding control region (D-loop). These genes except ND6 and 8 tRNA genes were encoded on the H-strand. The phylogenetic analysis exhibited that our sequence formed a sister branch with P. cinereal and P. nemaeus of genus Pygathrix, which showed a closer genetic relationship of the three species. These information contribute to molecular, phylogenetic studies and genetic diversity conservation for this species.
Black-shanked douc langur (Pygathrix nigripes), taxonomically affiliated to the subfamily Colobinae in the family Cercopithecidae (Hasegawa et al. Citation2002; Duc et al. Citation2009), mainly inhabited evergreen/semi-evergreen forests and rainforests in eastern Cambodia and southeast Vietnam with a highly narrow habitat range (Thuc et al. Citation2005; Bett et al. Citation2012). Owing to illegal hunting for traditional ‘medicine’, pet trade and natural habitat destruction, the wild populations of P. nigripes have reduced dramatically (Nadler Citation2008), and it was classified as Endangered (EN) in the International Union for Conservation of Nature (IUCN Citation2007) Red List of Threatened Species (Rawson et al. 2008). In the present paper, we first determined and characterized the complete mitogenome of P. nigripes to contribute to its molecular, phylogenetic studies and genetic diversity conservation.
The muscle tissue of the black-shanked douc langur was collected from a natural death individual in Nanning, China (22°50′N, 108°15′E), and it was returned to the zoology lab and stored at −80 °C in Sichuan Agricultural University. Genomic DNA was extracted by phenol-chloroform extraction method (Sambrook et al. Citation1990). Sequences’ homology was analyzed using Megalign of the DNAStar (Swayne et al. Citation2015). Nucleotide variation loci were calculated by DnaSP v5 (Librado and Rozas Citation2009). Phylogenetic tree was constructed using Mega 7.0 (Kumar et al. Citation2016).
The complete mitogenome sequence of P. nigripes (GenBank accession number MH064177) was 16,534 bp, including 13 PCGs, two rRNA genes (12S rRNA and 16S rRNA), 22 tRNA genes and one control region (D-loop). Genes encoding on the genome were similar among all primates (Roos Citation2018; Roos et al. Citation2018). The overall base composition is 32.4% A, 29% T, 25.9% C, 12.7% G, and the G + C content was 38.6%. Most PCGs started with ATG, while NADH2 and NADH3 genes initiated with ATT, NADH5 began with ATA, and NADH6 used TCT as start codon. All PCGs were terminated with typical TAA or TAG codons except for COX3, NADH3, NADH4, CYTB, which ended with incomplete stop codon AT- or T–, and NADH6 ended with CAT. These genes except ND6 and eight tRNA genes were encoded on the H-strand. The lengths of 12S rRNA and 16S rRNA were 1020 bp and 1086 bp, and separated by the tRNAVal gene. The D-loop was 1092 bp in length, and located between tRNAPro and tRNAPhe.
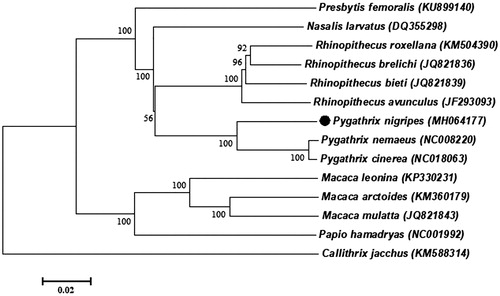
Phylogenetic analysis included mitogenome of P. nigripes and the other 12 species that are from Cercopithecinae and Colobinae, which belong to Primates, using Callithrix jacchus (Callithrix) as an outgroup (). The neighbour-joining (NJ) analysis exhibited that our sequence formed a sister branch with P. cinereal and P. nemaeus of genus Pygathrix, which showed a closer genetic relationship of the three species. Furthermore, the homology of 13PCGs between P. nigripes with P. nemaeus, P. cinereal is 92.75%, 92.51%, respectively, while 98.97% between P. cinereal and P. nemaeus, it suggested that P. cinereal and P. nemaeus have a closer genetic relationship. The result is in line with the topology of phylogenetic tree. This study provides new and comprehensive insight into the evolutionary and biogeographic history of this species, and contributes additional molecular information to species conservation.
Figure 1. NJ phylogenetic tree based on the complete mitochondrial genome of the P. nigripes and other 12 primates species sequences. Callithrix jacchus was served as an outgroup. Numbers at the branches indicated the bootstrapping values with 1000 replications. GenBank accession numbers were given in the parentheses. Filled circle represented a sequence from this study.
Disclosure statement
The authors declare that they have no conflict of interest and are alone responsible for the content and writing of the paper.
Additional information
Funding
References
- Bett NN, Blair ME, Sterling EJ. 2012. Ecological niche conservatism in doucs (genus pygathrix). Int J Primatol. 33:972–988.
- Duc HM, Baxter GS, Page MJ. 2009. Diet of Pygathrix nigripes, in southern Vietnam. Int J Primatol. 30:15–28.
- Hasegawa H, Murata K, Asakawa M. 2002. Enterobius (colobenterobius) pygatrichus sp. n. (nematoda: oxyuridae) collected from a golden monkey Pygathrix roxellana (milne-edwards, 1870) (primates: cercopithecidae: colobinae). Compar Parasitol. 69:62–65.
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451.
- Nadler T. 2008. Color variation in black-shanked douc langurs (Pygathrix nigripes), and some behavioural observations. Vietnam J Primatol. 2:71–76.
- Roos C. 2018. Complete mitochondrial genome of a toque macaque (macaca sinica). J Mitochondrial DNA Part B. 3:182–183.
- Roos C, Chuma IS, Collins DA, Knauf S, Zinner D. 2018. Complete mitochondrial genome of an olive baboon (papio anubis) from Gombe National Park, Tanzania. J Mitochondrial DNA Part B. 3:177–178.
- Sambrook J, Fritsch EF, Maniatis T. 1990. Molecular cloning: a laboratory manual. Anal Biochemist. 186:182–183.
- Swayne DE, Suarez DL, Spackman E, Jadhao S, Dauphin G, Kim-Torchetti M, McGrane J, Weaver J, Daniels P, Wong F, et al. 2015. Antibody titer has positive predictive value for vaccine protection against challenge with natural antigenic-drift variants of h5n1 high-pathogenicity avian influenza viruses from Indonesia. J Virol. 89:3746–3762.
- Thuc PD, Covert H, Polet G, Becker I, Mui TV. 2005. New survey data on pygathrix nigripes, the black-shanked douc langur, from cat tien national park, vietnam. Am J Phys Anthropol. 40:165–166.
- The IUCN Red List of Threatened Species. 2017. Version 2017-3; [accessed 2018 June 14]. www.iucnredlist.org.
