Abstract
The Siberian Shrew, Crocidura sibirica, is distributed in northwest China and extended to Mongolia and central Asia. It has been listed as Near Threatened (NT) species recently by the red list of China’s vertebrates. Firstly, we obtained complete mitochondrial genome of Crocidura sibirica. In this study, the mitogenome sequence is 17,192 bp in length, containing 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, 1 replication origin (OL) and 1 control region. This data provides basic data to application in conservation genetics and evolution for this Near Threatened species.
Introduction
The Siberian Shrew Crocidura sibirica Dukelsky, 1930 belongs to the family Soricidae. This species distributes in northwest China and extends to Mongolia and central Asia. It has a rich habitat type such as mountain coniferous forest, the mountain valley and lakeshore swamp (Smith et al. Citation2009). The conservation status of this species has been classified as Least Concern (LC) since 1996 in IUCN. In China, Crocidura sibirica is regarded as a pest species and may consequently persecuted. This species has been considered as Near Threatened (NT) (Jiang et al. Citation2016). However, the mitochondrial (mt) genome sequence of Crocidura sibirica has not been described. In this study, we assembled and characterized the complete mitochondrial genome sequence of Crocidura sibirica. This data provides basic data to application in conservation genetics and evolution for this Near Threatened species.
A specimen of Crocidura sibirica was collected from Aletai, XinJiang Province (Latitude: 47.357°N, Longitude: 88.016°E), and maintained the specimen and its DNA in Animal Herbarium of Sichuan Normal University (Accession number: SQ64), Chengdu. Total genomic DNA was extracted from muscle tissue using the DNA extraction kit (Aidlab Biotech, Beijing, China). The mitochondrial genomes of Crocidura russula was used to design primers for polymerase chain reaction (PCR) and as template for gene annotation.
The complete mitochondrial genome of Crocidura sibirica was 17,192 bp in length. It comprises of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, 1 replication origin (OL) and 1 control region. Its gene order is identical to other insectivores (Chen et al. Citation2015; Kim et al. Citation2013; Nikaido et al. Citation2001). The gene order and gene position of the annular mitochondria have been listed and confirmed. The overall base composition of the complete genome was as follows: A (32.6%), T (32.7%), C (21.5%), and G (13.2%), with an A + T-rich pattern of the vertebrate mitochondrial genomes.
As in other Insectivores, 12 protein-coding genes were encoded on the H-strand, with only ND6 genes encoded on the L-strand. A majority of the tRNA genes are located on the heavy strand expect for tRNAIle/tRNAAla/tRNAAsn/tRNACys/tRNATyr/tRNASer/tRNAGlu/tRNAPro in the light strand. Total length of the 13 protein-coding genes is 11,718 bp. All of the protein-coding genes initiate with an ATG start codon except for ND2, ND3, and ND5, which begin with ATT, ATT and TTA, respectively. These protein-coding genes were terminated with TAG, TAA, TAT, TGC, CAT, and AGA. Overlap and noncoding bases in some genes are also observed.
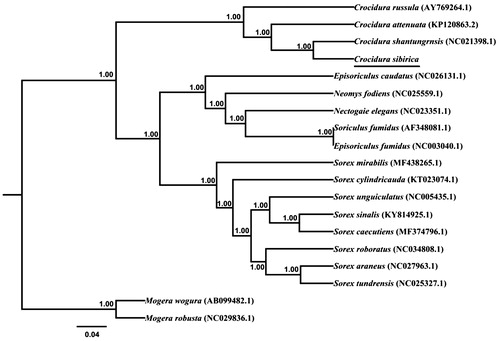
The phylogenetic relationship for Crocidura sibirica was examined with those of 18 Soricomorpha species previously reported in GenBank database. Phylogenetic trees were derived from the concatenated sequence of 12 protein-coding genes by employing Bayesian inference (BI) which was performed using BEAST v1.7.5 (Drummond et al. Citation2012). The best-fit TIM3 + I + G model was selected in JModelTest 0.1 (Darriba et al. Citation2012). Tree constructed using Bayesian is showed in . The three Crocidura species formed a solid monophyletic group, well supported by a Bayesian posterior probability of 1.00. Bayesian analyses also suggested that Crocidura sibirica was a sister relationship to Crocidura shantungensis (pp = 1.00).
Figure 1. Phylogenetic tree derived from 12 protein-coding gene sequences from 19 mitochondrial genomes of Soricomorpha using BI analysis. Numbers by the nodes indicate Bayesian posterior probabilities.
The present study accumulates mitochondrial genome data of the Siberian Shrew, Crocidura sibirica, may help us apply in conservation genetics and evolution for this Near Threatened species.
Acknowledgments
We are very grateful to Rui Liao from Sichuan Academy of Forestry for providing sample for this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen S, Tu F, Zhang X, Li W, Chen G, Zong H, Wang Q. 2015. The complete mitogenome of Stripe-Backed Shrew, Sorex cylindricauda (Soricidae). Mitochondr DNA. 26:477–478.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Rambaut A: Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Jiang Z, Jiang J, Wang Y, Zhang E, Zhang Y, Li L, Xie F, Cai B, Cao L, Zheng G, et al. 2016. Red list of China’s vertebrate. Biodivers Sci. 24:500–551.
- Kim HR, Park JK, Cho JY, Park YC. 2013. Complete mitochondrial genome of an Asian Lesser White-toothed Shrew, Crocidura shantungensis (Soricidae). Mitochondr DNA. 24:202–204.
- Nikaido M, Kawai K, Cao Y, Harada M, Tomita S, Okada N, Hasegawa M. 2001. Maximum likelihood analysis of the complete mitochondrial genomes of eutherians and a reevaluation of the phylogeny of bats and insectivores. J Mol Evol. 53:508–516.
- Smith AT, Xie Y, Hoffmann RS, Lunde D, MacKinnon J, Wilson DE, Wozencraft WC. 2009 A guide to the mammals of China. Changsha: Hunan Education Publish House.
