Abstract
Aspergillus egyptiacus is an important fungus during the fermentation process of dark tea. However, there is little study on genetic information about A. egyptiacus. In this study, A. egyptiacus was is isolated from dark tea and first sequenced. The annotation and analysis also have been done for the mitochondrial genome. The length of A. egyptiacus mitochondrial genome is 66,564 bp and include 41 genes. Similar to most fungus, 15 protein-coding genes (PCGs), 24 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and one non-coding control region (D-loop) were identified in the strand of mitochondrial genome. The GC content is 26.55% and the reconstructed phylogenetic supported the placement of A. egyptiacus in Eurotiomycetes clade. The mitochondrial genome information also may supply references for utilization of A. egyptiacus.
Aspergillus egyptiacus was first described by Moubasher and Moustafa in 1972 and was included in the A. nidulans group by most scientists (Moubasher and Moustafa Citation1972, Christensen and Raper Citation1978). A. egyptiacus could be used in many fields basing on its function, such as progesterone transformation (Ismail and Zohri Citation1994). As the unique microbial fermented tea, dark tea is fermented with microorganism during the manufacturing process. The research focus on the microorganism community is increasing in recent year for the health benefit value of dark tea. A. egyptiacus have been found during the fermentation process and showed playing an important role for quality determination(Guan et al. Citation2010, Li et al. Citation2017). Not according with the value of the important resource microorganism, there is little study on genetic information about A. egyptiacus.
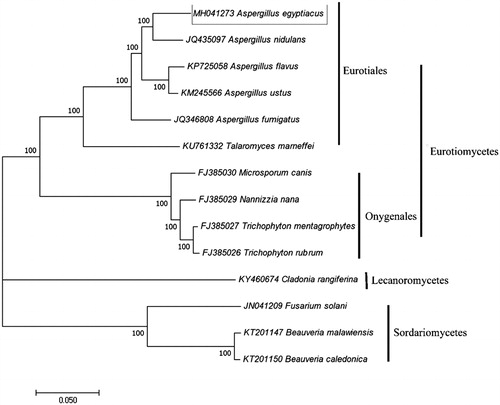
A. egyptiacus QA was isolated from dark tea in the Hunan Provincial Key Lab of Dark Tea and Jin-hua, Yiyang, China. The fungus was cultured in liquid medium (malt extract 20 g, yeast extract powder 20 g, sucrose 30 g, and water 1000 mL) with shaking at 180 rpm at 28 °C for 7 days. Mycelia were collected by filtration on Waterman paper and placed in a mortar, liquid nitrogen was added, and the samples were crushed using a pestle. Genomic DNA from was extracted using the CTAB method and sequences were obtained and assembled together using the Illumina HISeq2500 platform (Illumina, SanDiego, CA) with paired-end reads of 100 bp (Rogers and Bendich Citation1994). Adapters and low-quality reads were removed using the NGS QC Toolkit (Patel and Jain Citation2012). The circular was generated and genome was annotated as Wu Liang’s address (Liang et al. Citation2018). The sequence information of A. egyptiacus mitochondrial genome was submitted with the accession number MH041273. Then, maximum likelihood method was employed to constructed phylogenetic tree which includes 6 Eurotiales, 4 Onygenales, 3 Hypocreales and 1 Lecanorales species respectively using MEGA7.0 software with 1000 bootstrap replication (Kumar et al. Citation2016).
The length of A. egyptiacus mitochondrial genome is 66,564 bp and include 41 genes. Similar to most fungus, 15 protein-coding genes (PCGs), 24 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and one non-coding control region (D-loop) were identified in the strand of mitochondrial genome. The GC content is 26.55%, with composition of A, 38.24%, T, 35.21%, C, 11.52%, and G, 15.04% respectively. The reconstructed phylogenetic tree was clustered into three clades and the results supported the placement of A. egyptiacus in Eurotiomycetes clade (). The research confirms that A. egyptiacus has a close relationship to Aspergillus in molecular review. The mitochondrial genome information also may supply references for utilization of A. egyptiacus during dark tea fermentation process or other field.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Christensen M, Raper KB. 1978. Synoptic key to Aspergillus nidulans group species and related Emericella species. T British Mycol Soc. 71:177–191.
- Guan ML, Liu SC, Huang JA, Liu ZH, Jiang JL. 2010. Effect of fluorine on the growth of Eurotium Cristatum and Aspergillus niger fungi isolated from dark tea. J Tea Sci. 30:157–162.
- Ismail MA, Zohri AA. 1994. Confirmation of the relationships of Aspergillus egyptiacus and Emericella nidulans using progesterone transformation. Lett Appl Microbiol. 18:130–131.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li Q, Huang J, Li Y, Zhang Y, Luo Y, Chen Y, Lin H, Wang K, Liu Z. 2017. Fungal community succession and major components change during manufacturing process of Fu brick tea. Sci Rep. 7:6947.
- Liang W, Yunlin Z, Zhenggang X, Tian H, Libo Z, Shiquan L. 2018. The complete mitochondrial genome and phylogeny of Geospiza magnirostris (Passeriformes: Thraupidae). Conserv Gen Res. 2:1–3.
- Moubasher A.H, Moustafa A.F. 1972. Aspergillus egyptiacus sp. nov. Egypt J Bot. 153–154.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. Plos One. 7:e30619
- Rogers SO, AJ. Bendich 1994. Extraction of total cellular DNA from plants, algae and fungi. Netherlands: Springer.