Abstract
The grasshopper, Catantops pinguis Stål (Orthoptera: Catantopidae), is a worldwide pest in agriculture. We have obtained the whole mitochondrial genome of C. pinguis (GenBank accession No. MH671309). The entire mt genome is 15,604 bp long with an A + T content of 73.58%. This grasshopper mt genome encodes all the 37 genes that are typically found in animal mt genomes, consists of 13 protein-coding genes, 2 ribosomal RNA, and 22 transfer RNA genes. The gene order is consistent with other sequenced mt genome of grasshoppers. The A + T-rich region of this genome is 754 bp long with the A + T content of 87.27%, and located between the rrnS and trnI genes. Phylogenetic analysis was performed using 13 protein-coding genes with 11 grasshoppers of the family Catantopidae and it was seen that C. pinguis is closely related to Xenocatantops brachycerus.
The grasshopper, Catantops pinguis Stål (Orthoptera: Catantopidae), is a worldwide pest in agriculture. It is common in Africa and south and east Asia. Catantops pinguis causes considerable damage to the crops, such as maize, beans, and tea, because of its complex feeding habits (Zheng et al. Citation2013). In this study, C. pinguis were obtained from a tea plantation of Yongchuan in Chongqing province of China in May 2018. The specimens were stored in the insect specimen room of Tea Research Institute of Chongqing Academy of Agricultural Science with an accession number CQNKY-OR-01-01-01.
The complete mitochondrial genome of C. pinguis is a typical closed-circular DNA molecule with 15,604 bp in length, and the annotated sequence file was submitted to NCBI (GenBank accession No. MH671309). The total nucleotide composition of the major strand of the mt genome as follows: A = 42.60% (6,648), C = 15.41% (2,405), G = 11.03% (1,721), and T = 30.95% (4,830), with a total A + T content of 73.58% that is heavily biased toward A and T nucleotides. AT-skew and GC-skew of the whole J-strand of C. pinguis is 0.158 and −0.166, respectively. The mt genome of C. pinguis encodes all the 37 genes typically present in animal mt genomes, consisting of 13 protein-coding genes (PCG), two ribosomal RNA genes (rRNA) and 22 transfer RNA genes (tRNA). The mt genome of C. pinguis is compact, with total 39 bp overlapping at 9 gene junctions. Meanwhile, the mt genome has 106 bp intergenic sequence without the putative A + T-rich region. The A + T-rich region of this genome is 754 bp long with the A + T content of 87.27%, and located between the rrnS and trnI genes.
All of the 22 tRNA genes, usually found in insects mitochondrion, have been identified in C. pinguis. The gene size of tRNAs range from 64 bp (trnC) to 71 bp (trnV and trnK), and A + T content ranges from 63.08% (trnP) to 89.39% (trnE). Like most insects, 21 tRNA genes have typical cloverleaf shaped secondary structure and trnS1 lacks the dihydrouridine (DHU) arm (Chen et al. Citation2016; Wang et al. Citation2014). The two ribosomal RNA genes have been identified on the N-strand in the mt genome: the rrnL gene locates between trnL1 and trnV, and the rrnS gene between the trnV and the A + T-rich region. The length of rrnL and rrnS is 1,318 bp and 795 bp, and their A + T content is 85.95% and 73.46%, respectively.
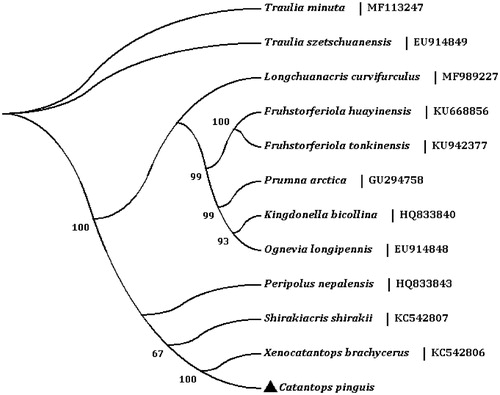
In the mt genome of C. pinguis, the total length of all 13 protein-coding genes is 11,188 bp, which accounts for 71.70% of the total genome. The A + T content of the 13 PCGs ranges from 65.53% (cox3) to 81.03% (nad6). All of the 13 PCGs initiate translation using ATN codons, and are terminated with TAA/TAG codons except cox1 with the incomplete T codon. The incomplete stop codon T is commonly reported and could produce functional stop codons in polycistronic transcription cleavage and polyadenylation mechanisms (Boore Citation2001; Ojala et al. Citation1981). We analyzed the amino acid sequences of 13 PCGs with maximum likelihood (ML) method to learn the phylogenetic relationship of C. pinguis with other grasshoppers in family Catantopidae. The phylogenetic tree shows that C. pinguis is closely related to Xenocatantops brachycerus ().
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18:1413–1416.
- Chen SC, Wang XQ, Wang JJ, Hu X, Peng P. 2016. The complete mitochondrial genome of a tea pest looper, Buzura suppressaria (Lepidoptera: Geometridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3153–3154.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Wang Y, Li H, Wang P, Song F, Cai W. 2014. Comparative mitogenomics of plant bugs (Hemiptera: Miridae): identifying the AGG codon reassignments between serine and lysine. PLoS ONE. 9:e101375.
- Zheng H, Li L, Xu Q, Zou Q, Tang B, Wang S. 2013. Gene cloning and expression patterns of two prophenoloxidases from Catantops pinguis (Orthoptera: Catantopidae). Bull. Entomol. Res. 103:393–405.