Abstract
The whole chloroplast genome (cp DNA) sequence of Caulerpa lentillifera J. Agardh has been characterized from Illumina pair-end sequencing. The circular cpDNA was 119,402 bp in length, containing 122 genes, which included 91 protein-coding genes, 28 tRNA genes, and 3 ribosomal RNA genes (four rRNA species). The overall AT content of C. lentillifera cpDNA is 67.4%. The 48 genes phylogenetic analysis suggested that C. lentillifera formed a monophyletic clade with congeneric C. racemosa.
Caulerpa lentillifera J. Agardh is widely distributed in the tropic Indo-Pacific (Phillips et al. Citation1999; Schils and Coppejans Citation2003; Kazi et al. Citation2013). As an edible alga called “sea grape”, C. lentillifera contains multiple essential amino acids with low-level total lipid (Niwano et al. Citation2009). The cultivations of this species are conducted in the Philippines, Malaysia, Japan, Vietnam and China (Zemke-White and Ohno Citation1999; Kurashima et al. Citation2003; Shi Citation2008). In the present study, we assembled and characterized the complete chloroplast genome sequence of C. lentillifera (MG753774) based on the Illumina pair-end sequencing data.
The fresh thalli of C. lentillifera were collected from Lingtou sea area (Hainan, China; 18°68′N, 108°70′E), and were used for the total genomic DNA extraction with the modified CTAB method (Doyle and Doyle Citation1987). DNA samples and voucher specimens were deposited in the Marine Biological Museum of the Chinese Academy of Sciences (MBMCAS). The whole-genome sequencing was conducted with 125 bp pair-end reads on the Illumina Hiseq Platform (Illumina, San Diego, CA). In all, 686 M raw reads were obtained, and after the quality-trimmed using the software CLC Genomics Workbench v7.5 (CLC bio, Aarhus, Denmark), the resultant 643 M clean reads were then used for the cpDNA assembly using the MITObim v1.7 (Hahn et al. Citation2013). The complete chloroplast genome sequence annotation of C. lentillifera was conducted by web server DOGMA (Wyman et al. Citation2004). Some tRNAs, rRNAs and coding sequences were further confirmed, and manually adjusted after BLAST searches. The complete chloroplast genome of C. lentillifera was 119,402 bp in length. This circular chloroplast genome contains 91 protein-coding genes, 28 tRNA genes, and 3 rRNA genes. Among the tRNA genes, the tRNAMet have three copies. The content of A, C, G, and T in the chloroplast genome was 32.9%, 16.0%, 16.6%, and 34.5%, respectively. Besides, the GC content of the whole chloroplast genome was 32.6%.
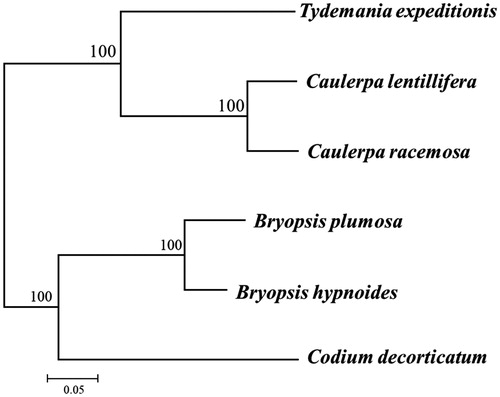
A phylogenetic analysis was carried out based on concatenated dataset of 48 genes (atpA, atpB, atpE, atpH, atpI, clpP, infA, petA, petB, petG, psaA, psaB, psaC, psaJ, psbA, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbN, psbT, rbcL, rpl2, rpl5, rpl14, rpl16, rpl20, rpl23, rpl36, rps3, rps4, rps7, rps8, rps9, rps11, rps12, rps14, rps18, rps19, tufA, ycf3, ycf4, and ycf12) with a GTR + G model implemented in RAxML8.1 (Stamatakis Citation2014). The topology of the phylogenetic tree revealed that C. lentillifera clustered with congeneric C. racemosa (KT946602) in a monophyletic clade (). The newly characterized C. lentillifera chloroplast genome will provide essential data for further studies on phylogeny and evolution of Bryopsidales and provide valuable insight conservation and restoration efforts for this economic important alga.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41: e129–e129.
- Kazi MA, Reddy CR, Jha B. 2013. Molecular phylogeny and barcoding of Caulerpa (Bryopsidales) based on the tufA, rbcL, 18S rDNA and ITS rDNA genes. PLoS One. 8:e82438.
- Kurashima A, Serisawa Y, Kanbayashi T, Toma T, Yokohama Y. 2003. Characteristics in photosynthesis of Caulerpa lentillifera J. Agardh and C. racemosa (Forsskal) J. Agardh var. laete-virens (Montagne) Weber-van Bosse with reference to temperature and light intensity. Jpn J Phycol. 51:167–172.
- Niwano Y, Beppu F, Shimada T, Kyan R, Yasura K, Tamaki M, Nishino M, Midorikawa Y, Hamada H. 2009. Extensive screening for plant foodstuffs in Okinawa, Japan with anti-obese activity on adipocytes in vitro. Plant Foods Hum Nutr. 64: 6–10.
- Phillips J, Conacher C, Horrocks J. 1999. Marine macroalgae from the Gulf of Carpentaria, tropical northern Australia. Aust Syst Bot. 12: 449–478.
- Schils T, Coppejans E. 2003. Phytogeography of upwelling areas in the Arabian Sea. J Biogeography. 30:1339–1356.
- Shi JH. 2008. Field survey and culture studies of Caulerpa in Taiwan. Dissertation, National Sun Yat-sen University, Taipei.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zemke-White WL, Ohno M. 1999. World seaweed utilisation: An end-of-century summary. J App Phycol. 11:369–376.