Abstract
Hoya is the largest genus (about 350–450 species) within Apocynaceae. It is a subshrub or liana, epiphytic or epilithic. Most species grow in tropical and subtropical South and Southeast Asia. Here we report and characterize the complete plastid genome sequence of Hoya pottsii Traill and Hoya liangii Tsiang in an effort to provide genomic resources useful for promoting its systematics research. The complete plastome of H. pottsii is 161,565 bp in length, including two Inverted Repeat (IR) regions of 24,657 bp, a Large Single-Copy (LSC) region of 92,532 bp, and a Small Single-Copy (SSC) region of 19,719 bp. The plastome contains 115 genes, consisting of 81 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of H. pottsii is 62.40%. The complete plastome of H. liangii is 162,989 bp in length, including two IR regions of 24,841 bp, a LSC region of 93,292 bp, and a SSC region of 20,015 bp. The plastome contains 115 genes, consisting of 81 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. The overall A/T content in the plastome of H. pottsii is 62.30%. The complete plastome sequence of H. pottsii and H. liangii will provide a useful resource for the conservation genetics of the two species as well as for the phylogenetic studies of Hoya.
Introduction
Hoya is the largest genus (about 350–450 species) within Apocynaceae. It is a subshrub or liana, epiphytic or epilithic. Most species grow in tropical and subtropical South and Southeast Asia (Lamb and Rodda Citation2016). There are 38 species and one variety in China, and most of them are distributed in southwestern China (Zhang et al. Citation2015). The leaves of H. pottsii are used for the treatment of fractures and swellings and for draining off pus and promoting new growth. Consequently, the genetic and genomic information is urgently needed to promote its medicinal and systematics research of H. pottsii and H. liangii. Here, we report and characterize the complete plastome of H. liangii (GenBank accession number, MH678666, this study) and H. pottsii (MH678667, this study). This is the first report of a complete plastome for the genus Hoya.
In this study, H. pottsii and H. liangii were sampled from Luoyi village, Chengmai county in Hainan province of China (110.111°E, 19.897°N). The voucher specimens (H. pottsii, Wang et al., B265 and H.liangii, Wang et al., B266) were deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
The experiment procedure is as reported in Zhu et al. (Citation2018). Around 6 Gb clean data were assembled against the plastome of Cynanchum wilfordii (KT220733.1) (Jang et al. Citation2016) using MITObim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of C. wilfordii (KT220733.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastomes of H. pottsii and H. liangii were found to possess a total length 161,565 bp and 162,989 bp, respectively. They both have typical quadripartite structure of angiosperms, containing two Inverted Repeats (IRs) of 24,657 bp in H. pottsii and 24,841 bp in H. liangii, a Large Single-Copy (LSC) region of 92,532 bp (H. pottsii) and 93,292 bp (H. liangii), a Small Single-Copy (SSC) region of 19,719 bp (H. pottsii) and 20,015 bp (H. liangii). The plastome of H. pottsii and H. liangii contains 115 genes, consisting of 81 unique protein-coding genes (7 of which are duplicated in the IR), 30 unique tRNA genes (7 of which are duplicated in the IR), and 4 unique rRNA genes. Among these genes, 5 pseudogenes (rpoC1, accD, ycf2, ndhH, ycf1) in H. pottsii and 6 pseudogenes (rpoC2, accD, ycf2, ycf1, ndhH, ycf1) in H. liangii, 14 genes possessed a single intron and 3 genes (ycf3, clpP, rps12) had 2 introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of H. pottsii is 62.40%, which the corresponding value of the LSC, SSC, and IR region were 64.10%, 68.20%, and 56.80%, respectively. The overall A/T content in the plastome of H. liangii is 62.30%, which the corresponding value of the LSC, SSC, and IR region were 64.10%, 68.20%, and 56.80%, respectively.
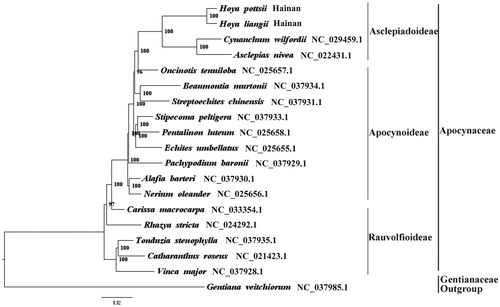
We used RAxML(Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 16 published complete plastome of Apocynaceae, using Gentiana veitchiorum (Gentianaceae) as outgroup. The phylogenetic analysis indicated that H. pottsii and H. liangii are close to C. wilfordii and Asclepias nivea within Apocynaceae respectively (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of H. pottsii and H. liangii will provide a useful resource for the conservation genetics of the two species as well as for the phylogenetic studies of Hoya.
Figure 1. The best ML phylogeny recovered from 19 complete plastome sequences by RAxML. Accession numbers: Hoya pottsii Traill (MH678667, this study), Hoya liangii Tsiang (MH678666, this study), Cynanchum wilfordii NC_029459.1, Asclepias nivea NC_022431.1, Oncinotis tenuiloba NC_025657.1, Beaumontia murtonii NC_037934.1, Streptoechites chinensis NC_037931.1, Stripecoma peltigera NC_037933.1, Pentalinon luteum NC_025658.1, Echites umbellatus NC_025655.1, Pachypodium baronii NC_037929.1, Alafia barteri NC_037930.1, Nerium oleander NC_025656.1, Carissa macrocarpa NC_033354.1, Rhazya stricta NC_024292.1, Tonduzia stenophylla NC_037935.1, Catharanthus roseus NC_021423.1, Vinca major NC_037928.1; outgroup: Gentiana veitchiorum NC_037985.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Jang W, Kim KY, Kim K, Lee SC, Park HS, Lee J, Seong RS, Shim YH, Sung SH, Yang TJ. 2016. The complete chloroplast genome sequence of Cynanchum auriculatum Royle ex Wight (Apocynaceae). Dna Seq. 27:4549–4550.
- Lamb A, Rodda M. 2016. A Guide to Hoyas of Borneo [M]. Borneo: Natural History Publications; p. 1–13.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang JF, Bai L, Xia NH, etal. 2015. Hoya yingjiangensis (Asclepiadoideae, Apocynaceae), a new campanulate flowered species from Yunnan, China. J. Phytotaxa. 219:283–288.
- Zhu ZX, Mu WX, Wang JH, Zhang JR, Zhao KK. Friedman CR, Wang HF. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia. Mitochondrial DNA Part B: Resources. 3(2):620–621.
