Abstract
Lonicera maackii is native to Northeastern Asia and considered as an invasive plant in North America. In this study, the complete chloroplast genome of L. maackii was obtained by high-throughput next-generation sequencing technology. The chloroplast genome was 155,318 bp in length and had typical quadripartite structures comprising a large single copy, a small single copy, and a pair of inverted repeat regions of 89,202, 18,680, and 23,718 bp long, respectively. The genome contained 80 protein-coding genes, four rRNA genes and 30 tRNA genes. Phylogenetic tree revealed that L. maackii was grouped with other Caprifoliaceae species, L. japonica, Patrinia saniculifolia, and Kolkwitzia amabilis.
Lonicera maackii (Amur honeysuckle) belongs to the Lonicera genus, Caprifoliaceae family. L. maackii is a woody perennial shrub which grows up to 5 m in height and sprouts earlier in spring. It is native to Northeastern Asia and has been widely used for ornamental purpose. In the late 1800s, L. maackii was imported to the Eastern United States (Forman Citation2011). However, L. maackii has been treated as an invasive plant due to its allelochemical that can suppress seed germination of other plants (Bauer et al. Citation2012). So far, genetic information of L. maackii was barely reported except for karyotype (Chen et al. Citation2017). The chloroplast genomes have been extensively used in understanding genetic diversity, authentication, and evolution in plants (Nguyen et al. Citation2018; Joh et al. Citation2017; Nguyen et al. Citation2017; Ye et al. Citation2014; Kim et al. Citation2017). In this study, we assembled and annotated the complete chloroplast genome sequence of L. maackii using next-generation sequencing technology.
The plant sample was collected from Medicinal Plant Garden, College of Pharmacy, Seoul National University, Koyang, Korea (37°42′45.3″N, 126°49′08.1″E), and its genomic DNA was isolated from fresh leaves using a modified cetyltrimethylammonium bromide protocol (Allen et al. Citation2006). The whole-genome sequencing was conducted using Illumina Miseq platform (Illumina, San Diego, CA). A total of 3.1 Gb reads were obtained and then de novo assembled using CLC genome assembler program (ver. 4.06 beta, CLC Inc, Aarhus, Denmark) as previously described (Kim et al. Citation2015). The chloroplast genome was initially annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseqapp.html) (Tillich et al. Citation2017) and manually corrected using Artemis annotation program (Rutherford et al. Citation2000) and BLASTN searches. The chloroplast genome sequence was submitted to the GenBank under accession number of MH028741.
The complete chloroplast genome of L. maackii had a total length of 155,318 bp, and showed a typical quadripartite structure with the large single copy, small single copy and a pair of inverted repeat regions; their sizes are 89,202; 18,680, and 23,718 bp, respectively. The chloroplast genome harbours 114 genes including 80 protein-coding genes, 4 ribosomal genes and 30 tRNA genes.
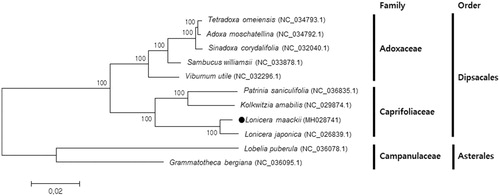
The aligned complete chloroplast genome sequences of L. maackii and 10 other species by MAFFT (Katoh and Standley Citation2013) were used for phylogenetic analysis. The neighbour-joining tree was constructed using MEGA6.0 with 1000 bootstrap replicates (Tamura et al. Citation2013). Phylogenetic analysis indicated L. maackii was clustered with three Caprifoliaceae species, and mostly close to another Lonicera species, L. japonica, which is widely used for traditional Chinese medicine (). The shorter branch length of L. maackii than L. japonica may indicate that L. maackii is closer to other Caprifoliaceae species. The complete chloroplast genome sequence of L. maackii can be utilized for DNA barcoding and evolutionary studies of the genus Lonicera.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320.
- Bauer JT, Shannon SM, Stoops RE, Reynolds HL. 2012. Context dependency of the allelopathic effects of Lonicera maackii on seed germination. Plant Ecol. 213:1907–1916.
- Chen J, Xia N, Wang X, Beeson RC, Chen J. 2017. Ploidy Level, Karyotype, and DNA Content in the Genus Lonicera. HortSci. 52:1680–1686.
- Forman T. 2011. Applying landscape ecology in biological conservation. Springer Science & Business Media, New York.
- Joh HJ, Kim N-H, Jayakodi M, Jang W, Park JY, Kim YC, In J-G, Yang T-J. 2017. Authentication of Golden-Berry P. ginseng Cultivar ‘Gumpoong’from a Landrace ‘Hwangsook’Based on pooling method using chloroplast-derived markers. Plant Breed Biotechnol. 5:16–24.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast ad ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kim K, Nguyen VB, Dong J, Wang Y, Park JY, Lee S-C, Yang T-J. 2017. Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species. Sci Rep. 7:4917.
- Nguyen VB, Park H-S, Lee S-C, Lee J, Park JY, Yang T-J. 2017. Authentication markers for five major Panax species developed via comparative analysis of complete chloroplast genome sequences. J Agric Food Chem. 65:6298–6306.
- Nguyen VB, Giang VNL, Waminal NE, Park H-S, Kim N-H, Jang W, Lee J, Yang T-J. 2018. Comprehensive comparative analysis of chloroplast genomes from seven Panax species and development of an authentication system based on species-unique SNP markers. J Ginseng Res. doi:10.1016/j.jgr.2018.06.003
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16:944–945.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Ye C-Y, Lin Z, Li G, Wang Y-Y, Qiu J, Fu F, Zhang H, Chen L, Ye S, Song W, et al. 2014. Echinochloa chloroplast genomes: insights into the evolution and taxonomic identification of two weedy species. PloS One. 9:e113657.