Abstract
Diospyros is the largest genus (about 485 species) in Ebenaceae. It is a deciduous or evergreen tree or shrub. It grows in pantropical and extending into temperate regions. Here, we report and characterize the complete plastid genome sequences of D. maclurei Merr. and D. hainanensis Merr. in an effort to provide genomic resources useful for promoting their systematics research and potential economic development. The complete plastome of D. maclurei is 157,946 bp in length, including two Inverted Repeat (IR) regions of 26,081 bp, a Large Single-Copy (LSC) region of 87,387 bp, and a Small Single-Copy (SSC) region of 18,397 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and four unique rRNA genes. The overall A/T content in the plastome of D. maclurei is 62.60%. The complete plastome of D. hainanensis is 157,999 bp in length, including two Inverted Repeat (IR) regions of 26,077 bp, a Large Single-Copy (LSC) region of 87,523 bp, and a Small Single-Copy (SSC) region of 18,322 bp. The plastome contains 114 genes, consisting of 80 unique protein-coding genes, 30 unique tRNA genes, and four unique rRNA genes. The overall A/T content in the plastome of D. hainanensis is 62.60%. The complete plastome sequences of D. Maclurei and D. hainanensis will provide a useful resource for the conservation genetics of the two species as well as for the phylogenetic studies of Diospyros.
Introduction
Diospyros maclurei and Diospyros hainanensis were sampled from Diaoluo Mountain (109.88°E, 18.67°N), which is a National Nature Reserve of Hainan, China. The voucher specimens (D. maclurei, Wang et al., B51 and D. hainanensis, Wang et al., B10) were deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China. The experiment procedure is as reported in Zhu et al. (Citation2018). Around 6 Gb clean data were assembled against the plastome of D. blancoi (NC033502.1) (Jo et al. Citation2016) using MITObim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of D. blancoi (NC033502.1) (Jo et al. Citation2016). The annotation was corrected with DOGMA (Wyman et al. Citation2004). The plastomes of D. maclurei and D. hainanensis were found to possess a total length 157,946 bp and 157,999 bp, respectively. They both have typical quadripartite structure of angiosperms, containing two Inverted Repeats (IRs) of 26,081 bp in D. maclurei and 26,077 bp in D. hainanensis, a Large Single-Copy (LSC) region of 87,387 bp (D. maclurei) and 87,523 bp (D. hainanensis), and a Small Single-Copy (SSC) region of 18,397 bp (D. maclurei) and 18,322 bp (D. hainanensis). Both plastomes contain 114 genes, consisting of 80 unique protein-coding genes (six of which are duplicated in the IR), 30 unique tRNA gene (seven of which are duplicated in the IR) and four unique rRNA genes (16S rRNA, 23S rRNA, 4.5S rRNA, and 5S rRNA). Among these genes, two pseudogenes [ycf1 (translation from 112,295 to 113,481) and ycf1 (translation from 133,039 to 127,419)] in D. maclurei and one pseudogene [ycf1 (translation from 112,427 to 113,613)] in D. hainanensis, 15 genes possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of D. maclurei and D. hainanensis is 62.60%, which the corresponding value of the LSC, SSC, and IR region were 64.60%, 69.20%, and 56.90%, respectively.
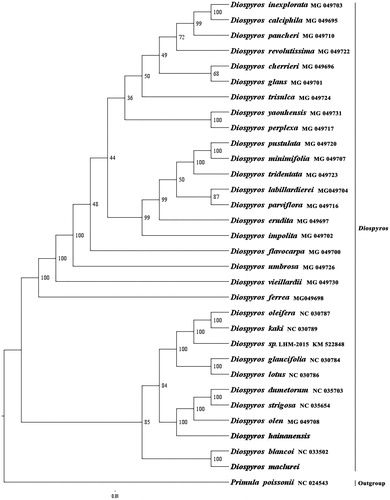
We used RAxML (Stamatakis, Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 29 published complete plastomes of Ebenaceae, using Primula poissonii (Sapotaceae) as outgroup. The phylogenetic analysis indicated that D. maclurei is close to D. blancoi, and D. hainanensis is close to the ancestor of D. olen, D. dumetorum, and D. strigosa within Diospyros (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequences of D. maclurei and D. hainanensis will provide a useful resource for the systematics and economic development of the two species as well as for the phylogenetic studies of Diospyros.
Figure 1. The best ML phylogeny recovered from 32 complete plastome sequences by RAxML. Accession numbers: Diospyros maclurei Merr. (This study, GenBank accession number: MH778101), Diospyros hainanensis Merr. (This study, GenBank accession number: MH778100), Diospyros minimifolia MG_049707, Diospyros pustulata MG_049720, Diospyros tridentata MG_049723, Diospyros labillardierei MG_049704, Diospyros parviflora MG_049716, Diospyros erudita MG_049697, Diospyros impolita MG_049702, Diospyros perplexa MG_049717, Diospyros yaouhensis MG_049731, Diospyros flavocarpa MG_049700, Diospyros umbrosa MG_049726, Diospyros calciphila MG_049695, Diospyros inexplorata MG_049703, Diospyros pancheri MG_049710, Diospyros revolutissima MG_049722, Diospyros glans MG_049701, Diospyros cherrieri MG_049696, Diospyros trisulca MG_049724, Diospyros vieillardii MG_049730, Diospyros ferrea MG_049698, Diospyros oleifera NC_030787, Diospyros kaki NC_ 030789, Diospyros sp. LHM-2015 KM_522848, Diospyros glaucifolia NC_ 030784, Diospyros lotus NC _030786, Diospyros dumetorum NC_ 035703, Diospyros strigosa NC_ 035654, Diospyros olen MG_049708, Diospyros blancoi NC _033502; outgroup: Primula poissonii NC_024543.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Jo S, Kim HW, Kim YK, Cheon SH, Kim KJ. 2016. The complete plastome sequence of Diospyros blancoi a. dc. (Ebenaceae). Mitochondrial DNA Part B. 1:690–692.
- Stamatakis A. 2006. RAxML-VI-HPC maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhu ZX, Mu WX, Wang JH, Zhang JR, Zhao KK, CR, Friedman, Wang H-F. 2018. Complete plastome sequence of Dracaena cambodiana (Asparagaceae): a species considered “Vulnerable” in Southeast Asia.
