Abstract
Wolfiporia cocos have been used as diuretic, sedative, and tonic medicine. Although its medicinal effects were reported, its genomic information is rare. In this paper, we report the complete mitochondrial genome of Wolfiporia cocos which is a 136,067 bp circular structure with 5 protein-coding genes, 25 tRNA genes, and 2 rRNA genes. Additional six protein-coding genes (cob, cox1, cox2, cox3, nad2, nad6) were truncated or pseudogenized by internal stop codons. Phylogenetic analysis of the mitochondrial genome of W. cocos showed the distinct relationship with other five mitochondrial genomes of Polyporales.
Keywords:
W. cocos is a wood decay fungus. It is a subterranean mushroom with no mushroom cap. Usually, its shape is similar to that of coconuts but is irregular. W. cocos are filled with white mycelium and the outside is covered with brown integument. It is used in traditional oriental medicine for its diuretic, sedative, and tonic effects (Ríos Citation2011). Its anti-cancer (Chen et al. Citation2009), anti-leukemia effect (Chen and Chang Citation2004), and anti-inflammatory action (Cuellar et al. Citation1997) were also reported.
In the taxonomical aspect, the ribosomal DNA of W. cocos has been studied (Jo et al. Citation2017), but, the mitochondrial genome of W. cocos has not yet been reported. In this paper, we report the complete mitochondrial genome sequence of W. cocos by assembly of Illumina whole genome sequence. The nucleotide sequence and annotation information have been deposited in GenBank with accession no. MH661258
Fresh samples of W. cocos strain PC1609 were provided by DONG-A PHARM (Seoul, South Korea), which were cultivated in the farm area located in Chungcheongnam-do, Geumsan-gun, South Korea. Total DNA was extracted from a single mushroom’s surface using CTAB method (Allen et al. Citation2006). The library preparation and sequencing was conducted with Illumina MiSeq platform following manufacturer’s instruction. After quality trimming, pair-end reads with Phred score 20 or more were assembled by a CLC genome assembler (ver 4.06 beta, CLC Inc, Rarhus, Denmark), following de-novo assembly of low coverage whole genome sequence (Kim et al. Citation2015). We used the Trametes cingulate sequence (GenBank Acc. NC_013933) as a reference genome for selecting representative contigs and joining them into a single draft sequence. For gene annotation, we used GeSeq (Tillich et al. Citation2017) program and confirm it with manual curation through the BLASTN search.
The complete mitochondrial genome of W. cocos is a circular form with 136,067 bp in length. Totally 389,000 of trimmed reads were aligned on the whole mitochondrial genome with the coverage of 712.25x. The mitogenome contained 5 protein-coding genes (atp6, atp8, nad3, nad4L, nad5), 25 tRNA genes, and 2 rRNA genes (rrn16, rrn23). In a comparison with the reference genome, 6 protein-coding genes (cob, cox1, cox2, cox3, nad2, nad6) were incomplete structure by truncation or became pseudogenes by presence of internal stop codons.
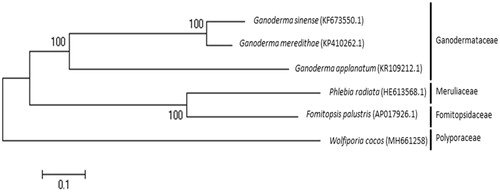
For phylogenetic analysis, we compared whole mitochondrial genome sequence of W. cocos with other five mitogenomes in Polyporales deposited in GenBank by a maximum likelihood analysis using MEGA 7.0 (Tamura and Nei Citation1993). W. cocos belonged in Polyporaceae is separated from other families in Polypolales. Ganodermataceae family made a separated clade while Phlebia radiate and Fomitopsis palustris were clustered into the same clade (). This new complete mitochondrial sequence of W. cocos will provide basic information for further genetic analysis and contribute for identification of this fungus.
Disclosure statement
The authors report that they have no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Allen G, Flores-Vergara M, Krasynanski S, Kumar S, Thompson W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1:2320.
- Chen X, Xu X, Zhang L, Zeng F. 2009. Chain conformation and anti-tumor activities of phosphorylated (1→ 3)-β-d-glucan from Poria cocos. Carb Pol. 78:581–587.
- Chen Y-Y, Chang H-M. 2004. Antiproliferative and differentiating effects of polysaccharide fraction from fu-ling (Poria cocos) on human leukemic U937 and HL-60 cells. Food Chem Toxicol. 42:759–769.
- Cuellar MJ, Giner RM, del Carmen RECIO M, JUST MJ, MANEZ S, RIOS JL. 1997. Effect of the basidiomycete Poria cocos on experimental dermatitis and other inflammatory conditions. Chem Pharm Bull. 45:492–494.
- Jo W-S, Lee S-H, Koo J, Ryu S, Kang M-G, Lim S-Y, Park S-C. 2017. Morphological characteristics of fruit bodies and basidiospores of Wolfiporia cocos. J. mushrooms 15:54–56.
- Kim K, Lee S-C, Lee J, Yu Y, Yang K, Choi B-S, Koh H-J, Waminal NE, Choi H-I, Kim N-H, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- Kim K, Lee S-C, Lee J, Lee HO, Joh HJ, Kim N-H, Park H-S, Yang T-J. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng Species. PLoS One. 10:e0117159.
- Ríos J-L. 2011. Chemical constituents and pharmacological properties of Poria cocos. Planta Medica. 77:681–691.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.