Abstract
Mazus pumilus (N. L. Burman) Steenis is the representative species of Mazus mainly distributed in China. Here, we report the complete chloroplast genome sequence of M. pumilus. The genome was 153,149 bp in length with 106 genes comprising 79 protein-coding genes, 23 tRNA genes, and 4 rRNA genes. The overall GC content of M. pumilus chloroplast genome was is 37.8%. ML phylogenomic analysis suggested that M. pumilus forms a monophyletic group with Lancea which shows a close relationship with the clade of Phrymaceae, Paulowniaceae, and Orobanchaceae.
Mazus Lour. (ca. 35 species) is mainly distributed in East Asia, Australia, and New Zealand, and about 25 species are found in China (Wu and Raven Citation1998). Mazus was firstly placed in Scrophulariaceae (Wettstein Citation1891). However, the systematic position of Mazus was altered by recent molecular-phylogenetic studies. Beardsley and Olmstead (Citation2002) found that Mazus and Lancea form a well-supported clade recognized as the subfamily Mazoideae belonging to the Phrymaceae. However, phylogenetic studies of Oxelman et al. (Citation2005), Albach et al. (Citation2009), Xia et al. (2009) and Schäferhoff et al. (Citation2010) confirmed that Mazus should be placed apart from the Phrymaceae. Based on the previous molecular-phylogenetic studies, Reveal (Citation2011) described a new family named Mazaceae which including Mazus, Lancea and Dodartia. Up to now, previous literature has not well revealed the phylogenetic relationship of Mazus and its related genus by different sequence fragments and need to be further elucidated.
In the present study, we report the completed chloroplast genomes of Mazus pumilus (N.L. Burman) Steenis which is the representative species of Mazus. M. pumilus was collected in Luoyang (112°26′45.2″E, 34°38′3.9″N, China) and the specimen was deposited in the Qinghai-Tibetan Plateau Museum of Biology (HNWP). The DNA was isolated from fresh leaves via the modified CTAB method (Doyle Citation1987). The complete chloroplast genome was sequenced at Novogene Biotech Co. (Tianjin, China) using the Illumina MiSeq platform. Genomic sequence was assembled with SOAPdenovo (Luo et al. Citation2012) and annotation was performed with CpGAVAS (Liu et al. Citation2012) by comparing with the previously reported chloroplast sequences of Lancea (Chi et al. Citation2018). The completed chloroplast genome sequences of M. pumilus together with 26 species from Lamiales and Lactuca sativa (outgroup) were aligned with MAFFT (Katoh and Standley 2013). Gblocks (Castresana Citation2000) was introduced to remove ambiguously aligned sites. A maximum likelihood (ML) analysis was implemented using RAxML-HPC2 on XSEDE based on the GTR + G + I nucleotide substitution model as recommended by jModelTest2 with 1000 replications.
The M. pumilus chloroplast genome (GenBank Accession No. MF593117) was 153,149 bp in length with a pair of inverted repeats (IR) regions (25,831 bp), a large single copy (LSC) region (84,034 bp), and a small single copy (SSC) region (17,453 bp). The GC content of the genome was 37.8%, and the GC contents of IR regions (43.1%) was higher than the LSC regions (35.7%) and SSC regions (32.1%). There were 106 predicted genes including 79 protein-coding genes, 23 tRNA genes, and 4 rRNA genes. Among the protein-coding genes, 63 were found in the LSC region, 11 were located in the SSC region, while ndhB, rpl2, rpl23, rps7, and ycf2 were duplicated in the IR regions.
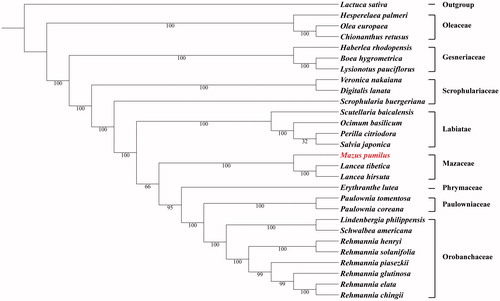
ML analysis showed that M. pumilus and Lancea species constituted one monophyletic group as Mazaceae (). Additionally, Mazaceae showed a close relationship with Phrymaceae, Paulowniaceae, and Orobanchaceae, rather than the Scrophulariaceae. This newly reported chloroplast data not only provided genomic information for Mazaceae but also revealed the phylogenetic relationships. These data will empower genetic engineering, conservation genetics and evolutionary studies involving this taxon.
Figure 1. Maximum likelihood phylogenetic tree based on 28 complete chloroplast genome sequences. The number on each node indicates the bootstrap value. Accession numbers: Boea hygrometrica JN107811, Chionanthus retusus KY582962, Digitalis lanata KY085895, Erythranthe lutea KU705476, Haberlea rhodopensis KX657870, Hesperelaea palmeri LN515489, Lactuca sativa AP007232, Lancea hirsuta MG551489, Lancea tibetica MF593117, Lindenbergia philippensis HG530133, Lysionotus pauciflorus KX752081, Ocimum basilicum KY623639, Olea europaea GU228899, Paulownia coreana KP718622, Paulownia tomentosa KP718624, Perilla citriodora KT220684, Rehmannia chingii KX426347, Rehmannia elata KX636161, Rehmannia glutinosa KX636157, Rehmannia henryi KX636158, Rehmannia piasezkii KX636160, Rehmannia solanifolia KX636159, Salvia japonica KY646163, Schwalbea americana HG738866, Scrophularia buergeriana KP718626, Scutellaria baicalensis KR233163, Veronica nakaiana KT633216.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Albach DC, Yan K, Jensen SR, Li HQ. 2009. Phylogenetic placement of Triaenophora (fromerly Scrophulariaceae) with some implications for the phylogeny of Lamiales. Taxon. 58:749–756.
- Beardsley PM, Olmstead RG. 2002. Redefining Phrymaceae: The placement of Mimulus, tribe Mimuleae, and Phryma. Am J Bot. 89:1093–1102.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Chi XF, Wang JL, Gao QB, Zhang FQ, Chen SL. 2018. The complete chloroplast genome of two Lancea species with comparative analysis. Molecules. 23:602.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and genbank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. Soapdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Oxelman B, Kornhall P, Olmstead RG, Bremer B. 2005. Further disintegration of Scrophulariaceae. Taxon. 54:411–425.
- Reveal JL. 2011. Summary of recent systems of angiosperm classification. Kew Bull. 66(1):5–48.
- Schäferhoff B, Fleischmann A, Fischer E, Albach DC, Borsch T, Heubl G, Müller KF. 2010. Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences. BMC Evol Biol. 10:352.
- Wettstein R. 1891. Nolanaceae, Solanaceae, Scrophulariaceae. Leipzig, Germany: Engelmann.
- Wu ZW, Raven PH. 1998. Flora of China: Vol. 18 Scrophulariaceae through Gesneriaceae. St. Louis, MO, USA: Missouri Botanical Garden.
- Xia Z, Wang YZ, Smith JF. 2009. Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s.L.) inferred from five gene regions. Am J Bot. 96:519–530.
