Abstract
Parasassafras confertiflorum (Meisn.) D.G. Long is the single tree species of the genus Parasassafras D.G. Long in the family Lauraceae. To better determine its phylogenetic location with respect to the related Lauraceae species, the complete chloroplast genome of P. confertiflorum was sequenced. The whole plastome is 152,555 bp in length, consisting of a pair of inverted repeat (IR) regions of 20,079 bp, one large single copy (LSC) region of 93,604 bp, and one small single copy (SSC) region of 18,793 bp. The genome contains 127 genes, including 83 protein-coding genes, 8 ribosomal RNA genes, and 36 transfer RNA genes. The overall GC content of the whole plastome is 39.1%. Further, maximum likelihoodphylogenetic analyses were conducted using 34 complete plastomes of the Lauraceae, which support close relationships between P. confertiflorum and Actinodaphne trichocarpa, Lindera benzoin, L. latifolia, L. metcalfiana, L. robusta, and Neolitsea sericea rather than Laurus nobilis or Sinosassafras flavinervium.
The genus Parasassafras D.G. Long in the family Lauraceae includes only one species Parasassafras confertiflorum (Meisn.) D.G. Long rarely distributed at high altitudes in Yunnan of SW China, Bhutan, India, and Myanmar (Huang and Henk Citation2008). For a better understanding of the relationships of P. confertiflorum and its related species, it is necessary to reconstruct a robust phylogenetic tree of the core Laureae group based on high-throughput sequencing approaches.
Young leaves of P. confertiflorum in Ximeng County (Yunnan, China; Long. 99.6687 E, Lat. 22.7615 N, 1788 m) were picked for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (Accession Number: XTBG-BRG-SY34285). The whole chloroplast genome was sequenced following Zhang et al. (Citation2016), and their 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. The contigs were aligned using the publicly available cp genome of Lindera glauca (GenBank accession number MF188124) and annotated in Geneious 4.8.
The plastome of P. confertiflorum (GenBank accession number MH729378.), with a length of 152,555 bp, was 65 bp and195 bp smaller than that of Litsea glutinosa (152,618 bp, KU382356) and Laurus nobilis (152,750 bp, KY085912). It was also 7 bp and 113 bp larger than that of the Lindera benzoin (152,478 bp, MH220730) and Neolitsea sericea (152,442 bp, MF939341) (Hinsinger and Strijk Citation2017; Song et al. Citation2017a; Zhao et al. Citation2018). The lengths of the inverted repeats (IRs), small single-copy (SSR) region, and large single-copy (LSC) region in the plastome of P. confertiflorum was 20,079 bp, 18,793 bp, and 93,604 bp, respectively. The G + C contents of the plastome is 39.1%.
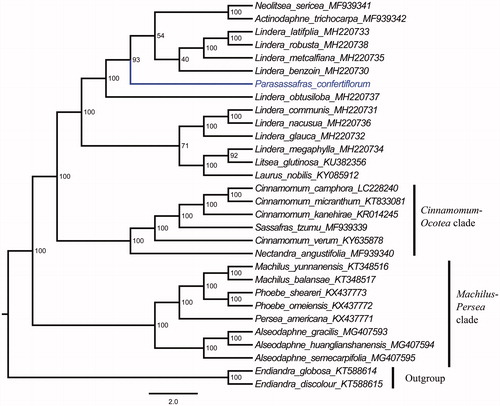
The complete plastid genome sequence of P. confertiflorum and the other sequenced Lauraceae taxa including Actinodaphne trichocarpa, Alseodaphne gracilis, A. huanglianshanensis, A. semecarpifolia, Cinnamomum camphora, C. kanehirae, C. micranthum, C. verum, Laurus nobilis, Lindera benzoin, L. communis, L. glauca, L. latifolia, L. megaphylla, L. metcalfiana, L. nacusua, L. obtusiloba, L. robusta, Litsea glutinosa, Machilus balansae, M. pauhoi, M. thunbergii, M. yunnanensis, Nectandra angustifolia, Neolitsea sericea, Persea americana, Phoebe chekiangensis, P. omeiensis, P. sheareri, P. zhennan, and Sassafras tzumu (Song et al. Citation2015, Citation2016, Citation2017a, Citation2017b, Citation2018; Hinsinger and Strijk Citation2017; Wu et al. Citation2017; Zhao et al. Citation2018), formed the base to perform a phylogenetic analysis, with Endiandra globosa and E. discolor as outgroup. A maximum likelihood analysis (Tamura et al. Citation2011) yielded a tree topology with 39–100% bootstrap (BS) values at each node. The topology divided the 32 Lauraceae taxa into three 100% supported clades (). The first divergent clade contained Alseodaphne, Machilus, Persea, and Phoebe taxa, the second Cinnamomum, Nectandra, and Sassafras taxa, and the third contained Actinodaphne, Laurus, Lindera, Litsea, Neolitsea species and P. confertiflorum. This phylogenetic tree reveals that Lindera is polyphyletic and there are close relationships between P. confertiflorum and Actinodaphne trichocarpa, Lindera benzoin, L. latifolia, L. metcalfiana, L. robusta, and Neolitsea sericea rather than Laurus nobilis or Sinosassafras flavinervium.
Acknowledgment
This work was supported by the Southeast Asia Biodiversity Research Institute [No. Y4ZK111B01] and “Light of West China” [No. Y7XB061B01] Programmes of CAS and the National Natural Science Foundation of China [No. 31600531].
Disclosure statement
The authors report no conflict of interest.
References
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36:715–722.
- Hinsinger DD, Strijk JS. 2017. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. 9:71–79.
- Huang PH, Henk VDW. 2008. Flora of China. In Wu ZY, Raven, PH, Hong DY, editors. Dipterocarpaceae, Vol. 13. St. Louis, MO: Science Press, Beijing, and Missouri Botanical Garden Press; p. 166.
- Song Y, Dong WP, Liu B, Xu C, Yao X, Gao J, Corlett RT. 2015. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Front Plant Sci. 6:662.
- Song Y, Yao X, Liu B, Tan YH, Corlett RT. 2018. Complete plastid genome sequences of three tropical Alseodaphne trees in the family Lauraceae. Holzforschung. 72:337–345.
- Song Y, Yao X, Tan YH, Gan Y, Corlett RT. 2016. Complete chloroplast genome sequence of the avocado: gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can J for Res. 46:1293–1301.
- Song Y, Yao X, Tan YH, Gan Y, Yang JB, Corlett RT. 2017b. Comparative analysis of complete chloroplast genome sequences of two subtropical trees, Phoebe sheareri and Phoebe omeiensis (Lauraceae). Tree Genet Genomes. 13:120.
- Song Y, Yu WB, Tan YH, Liu B, Yao X, Jin JJ, Padmanaba M, Yang JB, Corlett RT. 2017a. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the Magnoliids. Genome Biol. Evol. 9:2354–2364.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wu CC, Chu FH, Ho CK, Sung CH, Chang SH. 2017. Comparative analysis of the complete chloroplast genomic sequence and chemical components of Cinnamomum micranthum and Cinnamomum kanehirae. Holzforschung. 71:189–197.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Syst Evol. 54:219–227.
- Zhao ML, Song Y, Ni J, Yao X, Tan YH, Xu ZF. 2018. Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae). Sci Rep. 8:8844