Abstract
The complete mitochondrial genome of Paratrypauchen microcephalus (Gobiiformes: Oxudercidae) was completely sequenced by high throughput sequencing method. The complete mitochondrial genome was 16,552 bp in length, consisted of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, a putative control region (CR), and an origin of replication on the light-strand (OL). The base composition values for the mitochondrial genome were 29.1%, 27.6%, 15.7%, and 27.6% for A, C, G, and T, respectively. The gene arrangement is identical to those in typical fishes. Phylogenetic tree based on 13 protein-coding genes shows that P. microcephalus has a close phylogenetic relationship with genus Trypauchen and belongs to Oxudercidae.
The species comb goby, Paratrypauchen microcephalus (Gobiiformes: Oxudercidae), is distributed throughout the Indo-west Pacific, commonly inhabits soft mud bottoms of inshore and estuarine waters, and feeds on benthic invertebrates like crustaceans (Chen and Fang Citation1999; Murdy Citation2011). Studies on P. microcephalus were seldom and only research on length–weight relationship was performed (Yoon et al. Citation2013). Many species of Paratrypauchen have not been well recognized because of the absence of enough molecular information and clear phylogenetic relationship. As well known, mitochondrial DNA was proved effective in species identification and phylogenetic studies, so here we described the complete mitogenome of P. microcephalus for the first time and reconstructed the phylogenetic relationship of the relative species of Oxudercidae, and expecting for better understanding the systematic evolution of genus Paratrypauchen and further phylogenetic study of Gobiiformes.
The specimen was collected from Naozhou Island in Zhanjiang, China (geographic coordinate: N 20°53′20.11″, E 112°28′46.20″). The whole body specimen was preserved in ethanol and registered to the Marine Biodiversity Collection of South China Sea, Chinese Academy of Sciences, under the voucher number SW20181071705.
The complete mitochondrial genome of P. microcephalus was 16,552 bp in length (GenBank accession No. MH678617), containing 13 protein-coding genes, 22 tRNA genes, two rRNA genes, a putative control region (CR) as well as an origin of replication on the light-strand (OL). The gene arrangement is identical to those in typical fishes that most of these genes are encoded by the H-stand, except for ND6 and eight tRNA genes (Boore Citation1999; Liu et al. Citation2013; Gong et al. Citation2017). The 13 protein-coding genes encode 3803 amino acids in total and use the initiation codon ATG except COI using GTG and ATP6 using ATA. Most of them use TAA or TAG as the stop codon, while ATP6, COIII, and Cytb use an incomplete T or TA, COII uses an unusual AGA, and ND4 uses an unusual AGG. Overall base composition values for the mitochondrial genome were 29.1%, 27.6%, 15.7%, and 27.6% for A, C, G, and T, respectively, with slight AT-bias of 56.7%.
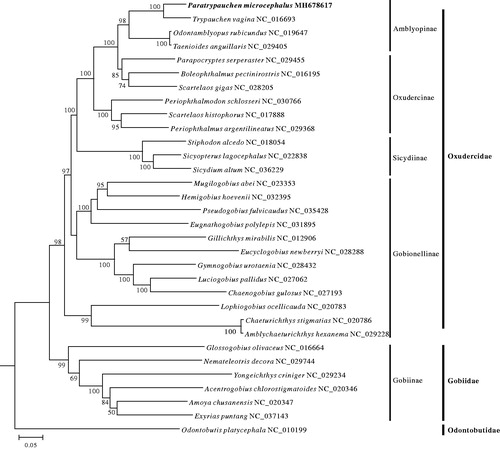
Based on first and second codon sequences of 13 protein-coding genes of each mitogenome from 31 species from Oxudercidae and Gobiidae, with Odontobutis platycephala as outgroup species, a maximum likelihood (ML) phylogeny tree was constructed by using MEGA6 (Tamura et al. Citation2013). In the ML phylogenetic tree, P. quadrilineatus and Trypauchen vagina first clustered together, and then grouped with Odontamblyopus rubicundus and Taenioides anguillaris in a strong support. This pylogenetic framework also supported the four subfamilies Amblyopinae, Oxudercinae, Sicydiinae and Gobionellinae together formed the Oxudercidae (Agorreta et al. Citation2013; Thacker et al. Citation2015; Nelson et al. Citation2016; Kuang et al. Citation2018) ().
Figure 1. Maximum-likelihood phylogenetic tree was constructed based on first and second codon sequences of 13 protein-coding genes of 32 species. The number at each node is the bootstrap probability (only show ≥50%). The number after the species name is the GenBank accession number, and the bold species is studied in this research.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Agorreta A, San Mauro D, Schliewen U, Van Tassell JL, Kovačić M, Zardoya R, Rüber L. 2013. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol Phylogenet Evol. 69:619–633.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Chen I-S, Fang L-S. 1999. The freshwater and estuarine fishes of Taiwan. Pingtung: National Museum of Marine Biology and Aquarium (In Chinese).
- Gong L, Du X, Lu ZM, Liu LQ. 2017. The complete mitochondrial genome characterization of Tridentiger obscurus (Gobiiformes: Gobiidae) and phylogenetic analyses of Gobionellinae. Mitochondrial DNA B-Resour. 2:662–663.
- Kuang T, Tornabene L, Li J, Jiang J, Chakrabarty P, Sparks JS, Naylor GJP, Li C. 2018. Phylogenomic analysis on the exceptionally diverse fish clade Gobioidei (Actinopterygii: Gobiiformes) and data-filtering based on molecular clocklikeness. Mol Phylogenetics Evol.128:192–202.
- Liu TX, Jin XX, Wang RX, Xu TJ. 2013. Complete sequence of the mitochondrial genome of Odontamblyopus rubicundus (Perciformes: Gobiidae): genome characterization and phylogenetic analysis. J Genet. 92:423–432.
- Murdy EO. 2011. Systematics of Amblyopinae. In: Patzner R, Van Tassell JL, Kovacic M, Kapoor BG, editors. The biology of gobies. Enfield, New Hampshire: Science Publishers; p. 107–118.
- Nelson JS, Grande TC, Wilson MV. 2016. Fishes of the world. New York: John Wiley & Sons.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Thacker CE, Satoh TP, Katayama E, Harrington RC, Eytan RI, Near TJ. 2015. Molecular phylogeny of Percomorpha resolves Trichonotus as the sister lineage to Gobioidei (Teleostei: Gobiiformes) and confirms the polyphyly of Trachinoidei. Mol Phylogenet Evol. 93:172–179.
- Yoon HS, An YK, Hwang JH, Lim HS, Lee WK, Han KH, Lee SH, Choi SD. 2013. Length–weight relationships for 14 fish species of the Suer River estuary in southern Korea. J Appl Ichthyol. 29:468–469.
