Abstract
The complete mitochondrial genome of the edible fungus Hypsizygus marmoreus was published in this paper. It was determined using Pacbio and Illumina sequencing. The complete mitochondrial DNA (mtDNA) is 106,417 bp in length with a GC content of 31.74%, which was the fourth large mitogenome in Agaricales. The circular mitogenome encoded 67 protein-coding genes and one ribosomal RNAs (rns). Among these genes, 13 conserved protein-coding genes were determined in the genome, including 6 subunits of NAD dehydrogenase (nad1-4, 4L and 6), three cytochrome oxidases (cox1-3), one apocytochrome b (cob) and three ATP synthases (atp6, apt 8 and apt 9). The phylogenic analysis confirmed that H. marmoreus (Lyophyllaceae) clustered together with Tricholoma matsutake (Tricholomataceae).
Introduction
Hypsizygus marmoreus (Peck) H. E. Bigelow, belonging to Agaricales, is a commercial edible mushroom. Because of its high nutritional and medicinal value, it is very popular in East Asian regions, including China and Japan (Wu et al. Citation2015). Some studies have shown that mushrooms are promising foods in improving human health and preventing diseases. In previous investigations, steroids, sphingolipids, proteins and polyisoprenepolyols were isolated from the fruiting bodies of this fungus (Akihisa et al. Citation2005; Jung et al. Citation2008; Krasnopolskaya et al. Citation2008; Lee et al. Citation2012). Some studies indicated that strains of H. marmoreus show abundant diversities in morphological and genetic characters. PCR-based molecular markers and inter-simple sequence repeat (ISSR) have been widely used in genetic studies of H. marmoreus (Wang et al. Citation2009; Lee et al. Citation2012; Qiu et al. Citation2013). However, there were no enough polymorphic sites for identifying different strains. The mitogenome reported here might provide important genetic information for further studies on phylogeny, strain conservation, identification or discrimination of the strains.
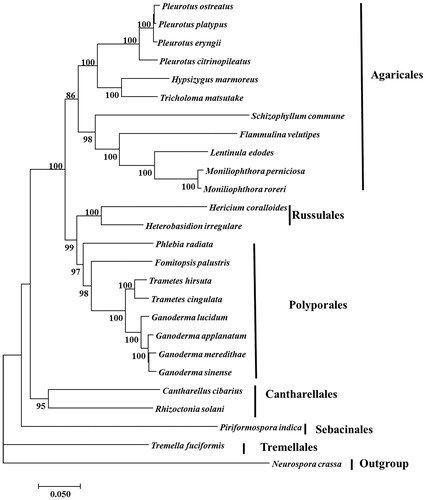
Monokaryoitc strain (F4 isolated from dikaryotic strain B5 deposited at Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences) isolation, genomic DNA extraction, strategies of sequencing (Illumina and Pacbio), sequencing processes, genome assembly and annotation, phylogenetic analysis were conducted according to the methods published previously (Yang et al. Citation2016a, Citation2016b, Citation2017; Wan et al. Citation2018). Assembly of a total of 258,199 Pacbio subreads (2,860,968,946 bp) and 67,421,744 high quality Illumina reads (9,955,446,419 bp) resulted in 106,417 bp of mitogenome which might be the fourth large mitogenome in Agaricales (Agaricus bisporus – 133 kb; Lentinula edodes – 116 kb; Moniliophthora perniciosa – 109 kb) (Férandon et al. Citation2013; Yang et al. Citation2017). The GC content was 31.74%. The circular mitogenome encoded 69 putative protein-coding genes and one ribosomal RNAs (rns). Thirteen conserved protein-coding genes encoded 6 subunits of NAD dehydrogenase (nad1-4, nad4L and nad 6), three cytochrome oxidases (cox1-3), one apocytochrome b (cob) and three ATP synthases (atp6, apt 8 and apt 9). Nine introns invaded into three genes, i.e. cob (1 intron), cox1 (7 introns) and cox2 (1 intron). These introns mainly belong to group IB. The 22 tRNA genes covered 19 standard amino acids, with the following three having two tRNAs each: two trnL (trnL-uaa and trnL-uag), two trnR (trnR-ucg and trnR-ucu) and two trnS (trnS-gcu and trnS-uga) while the remaining amino acids were each represented by only one tRNA gene. This genome could not encode Alanine. As shown in , the phylogenetic analysis confirmed that H. marmoreus (Lyophyllaceae) was a member of Agaricales and clustered together with Tricholoma matsutake (Tricholomataceae). It was in agreement with previous study, Tricholomatoid clade included four families, the Tricholomataceae s. str., Lyophyllaceae, Entolomataceae and Mycenaceae, and the Catathelasma clade (Matheny et al. Citation2006; Zhao et al. Citation2017). The evolutionarily relationship among Agaricales, Russulales, Polyporales, Cantharellales and Sebacinales was in agreement with results of previously study (Hibbett Citation2006; Garcia-sandoval et al. Citation2011; Zhao et al. Citation2017). The mitogenome of H. marmoreus would provide new insights into understanding the phylogeny and evolution of Lyophyllaceae and Agaricales.
Figure 1. Neighbour-joining tree of 26 species of Agaricomycotina conducted using MEGA 7.0 (Kumar et al. 2016) based on concatenated amino acid sequences of 13 mitochondrial protein-coding genes, including atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, and4, nad4L, nad5 and nad6. All the sequences were aligned using Clustal X (Thompson et al. 2010). The 25 other species used in this study were listed following: Cantharellus cibarius (NC_020368), Flammulina velutipes (NC_021373), Fomitopsis palustris (NC_034349), Hericium coralloides (NC_033903), Ganoderma applanatum (NC_027188), Ganoderma lucidum (NC_021750), Ganoderma meredithae (NC_026782), Ganoderma sinense (NC_022933), Heterobasidion irregulare (NC_024555), Lentinula edodes (NC_018365), Moniliophthora perniciosa (NC_005927), Moniliophthora roreri (NC_015400), Pleurotus citrinopileatus (NC_036998),Pleurotus ostreatus (NC_009905), Pleurotus platypus (NC_036999), Phlebia radiata (NC_020148), Rhizoctonia solani (HF546977), Schizophyllum commune (NC_003049), Serendipita indica (FQ859090), Trametes hirsuta (NC_037239), Tremella fuciformis(NC_036422), Trichosporon asahii var. asahii (MT: JH925097), Trametes cingulata (NC_013933) and Tricholoma matsutake (NC_028135). Neurospora crassa (NC_026614) was served as an outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches.
Data availability and accession numbers
This genome sequence has been deposited at NCBI (http://www.ncbi.nlm.nih.gov/) under the accession no. MH746465.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Akihisa T, Franzblau SG, Tokuda H, Tagata M, Ukiya M, Matsuzawa T, Metori K, Kimura Y, Suzuki T, Yasukawa K. 2005. Antitubercular activity and inhibitory effect on Epstein–Barr virus activation of sterols and polyisoprenepolyols from an edible mushroom, Hypsizigus marmoreus. Biol Pharm Bull. 28:1117–1119.
- Férandon C, Xu J, Barroso G. 2013. The 135 kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal Genet Biol. 55:85.
- Garcia-Sandoval R, Wang Z, Binder M, Hibbett DS. 2011. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia. 103:510–524.
- Hibbett DS. 2006. A phylogenetic overview of the Agaricomycotina. Mycologia. 98:917–925.
- Jung E-B, Jo J-H, Cho S-M. 2008. Nutritional component and anticancer properties of various extracts from haesongi mushroom Hypsizigus marmoreus. J Korean Society Food Sci. 37:1395–1400.
- Krasnopolskaya LM, Leontieva MI, Avtonomova AV, Isakova EB, Belitsky IV, Usov AI, Bukhman VM. 2008. Antitumor properties of submerged cultivated biomass and extracts of medicinal mushrooms of genus Hypsizygus singer Agaricomycetideae. Int J Med Mushrooms. 10:25–35.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. msw054.
- Lee CY, Park JE, Lee J, Kim JK, Ro HS. 2012. Development of new strains and related SCAR markers for an edible mushroom, Hypsizygus marmoreus. FEMS Microbiol Lett. 327:54–59.
- Lee CY, Song HS, Ro HS, Woo JR, You YH, Kim JG. 2012. Comparison of endo-, exo-cellular enzyme activity for new strains of Hypsizygus marmoreus. J Life Sci. 22:837–843.
- Matheny PB, Curtis JM, Hofstetter V, Aime MC, Moncalvo JM, Ge ZW, Slot JC, Ammirati JF, Baroni TJ, Bougher NL, et al. 2006. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia. 98:982–995.
- Qiu CS, Yan WJ, Peng L, Deng WQ, Song B, Li TH. 2013. Evaluation of growth characteristics and genetic diversity of commercial and stored lines of Hypsizygus marmoreus. Int J Agric Biol. 15:479–485.
- Thompson BJD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 2010. The CLUSTAL_X windows interface: exible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 2:4876–4882.
- Wan J, Li Y, Tang L, Wang H, Li Z, Zhou C, Bao D, Tan Q, Yang R. 2018. 3 complete mitochondrial genomes of straw-rotting edible fungus Volvariella volvacea using next generation sequencing. Mitochondrial DNA B. DOI: 10.1080/23802359.2018.1511849.
- Wang L, Hu X, Feng Z, Pan Y. 2009. Development of AFLP markers and phylogenetic analysis in Hypsizygus marmoreus. J Gen Appl Microbiol. 55:9–17.
- Wu F, Tang J, Pei F, Wang S, Chen G, Hu Q, Zhao L. 2015. The influence of four drying methods on nonvolatile taste components of white Hypsizygus marmoreus. Eur Food Res Technol. 240:823–830.
- Yang R, Li Y, Li C, Xu J, Bao D. 2016. The complete mitochondrial genome of the basidiomycete edible fungus Pleurotus eryngii. Mitochondrial DNA B. 1:772–774.
- Yang R, Li Y, Song X, Tang L, Li C, Tan Q, Bao D. 2017. The complete mitochondrial genome of t he widely cultivated edible fungus Lentinula edodes. Mitochondrial DNA B. 2:13–14.
- Yang RH, Li Y, Wáng Y, Wan JN, Zhou CL, Wāng Y, Gao YN, Mao WJ, Tang LH, Gong M, et al. 2016. The genome of Pleurotus eryngii provides insights into the mechanisms of wood decay. J Biotechnol. 239:65–67.
- Zhao RL, Li GJ, Sánchez-Ramírez S. 2017. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 84:1–32.
