Abstract
The mitogenome of Uroctea compactilis was 14,484 bp long. Total length of 13 protein-coding genes was 10,792 bp and nine of them were encoded on heavy strand (ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND6, and CYTB). Lengths of the two rRNA genes were 1061 bp (16S rRNA) and 696 bp (12S rRNA), respectively, and D-Loop was 688 bp long. The phylogenetic analysis, based on the completed mitogenome sequences of 11 species in eight families, revealed that the araneomorph spiders were divided into two groups of the orb-weaving spiders (Orbiculariae) and non-orb-weaving spiders (RTA clade and Eresoidea). Within the clade of non-orb-weaving spiders, U. compactilis (Oecobiidae) formed a sister clade to the RTA clade.
Spiders of Uroctea (Arachnida: Oecobiidae) is a genus of spiders that is found in Eurasia and Africa. There are two species of the genus Uroctea (U. compactilis and U. lesserti) recognized in Korea. Spiders of U. compactilis are distributed in southern part of Korea, while those of U. lesserti are found in northern part of the country (Kim et al. Citation2003).
We sequenced and characterized the complete mitogenome (MH752074) of an U. compactilis individual caught in southern part of South Korea. The voucher specimen was deposited in Genbank of the National Park Research Institute of Korea National Park Service. Genomic DNA extraction, PCR and gene annotation were conducted according to the previous studies (Kim et al. Citation2016a, Citation2016b). A previously published mitogenomes of Telamonia vlijmi (KJ598073) and Oxytate striatipes (KM507783) were used as references for primer design and gene annotation using Geneious version 8.0.5 (Auckland, New Zealand) (Kearse et al. Citation2012).
The mitogenome of U. compactilis was 14,484 bp long. Total length of 13 protein-coding genes was 10,792 bp and nine of them were encoded on heavy strand (ND2, COX1, COX2, ATP8, ATP6, COX3, ND3, ND6, and CYTB). Lengths of the two rRNA genes were 1061 bp (16S rRNA) and 696 bp (12S rRNA), respectively, and D-Loop was 688 bp long. In U. compactilis mitogenome, six kinds of initiation codon were found (ATA, ATC, ATG, ATT, TTA, and TTG). The start codon ATA is most common, which is found in six protein-coding genes (ATP8, ATP6, ND5, ND4, ND6, and CYTB), while the start codons ATC (ND3), ATG (COX3), TTA(COX1), and TTG (COX2) were only once used. The mitogenome of U. compactilis uses three kinds of stop codon (TAA, TAG, and T–) with TAA as most common stop codon. The stop codon T– was only once used in ND4L.
Higher phylogenetic relationship of spiders was inferred based on the completed mitogenomes of 11 species in eight spider families (). The phylogenetic tree was constructed based on the Kimura two-parameter model (Kimura Citation1980) using neighbour-joining (NJ) method (Saitou and Nei Citation1987) implemented in PAUP version 4.0B4a software (Sinauer Associates, Sunderland, MA) (Swofford Citation2002). Branch nodes of the tree were supported with 1000 bootstrap replicates. Ixodes pavlovskyi (NC_023831) (Acari) was used as outgroup.
Figure 1. Phylogenetic relationship of Uroctea compactilis and other spiders inferred based on complete mitogenome sequences of 11 species in eight spider families. The tree was generated under the Kimura’s two-parameter model using the neighbour-joining method implemented in PAUP version 4.0.b software. The robustness of the tree was tested with 1000 bootstrap. The numbers on the branches indicate bootstrap values. The mitogenome of U. compactilis in bold was determined and characterized in this study.
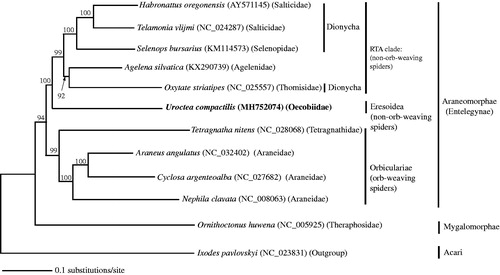
The phylogenetic analysis revealed that the two infraorders of Araneomorphae and Mygalomorphae (tarantulas and their close kin) were well separated. The araneomorph spiders were divided into two groups of the orb-weaving spiders (Orbiculariae) and non-orb-weaving spiders (RTA clade and Eresoidea). The RTA clade is a clade of araneomorph spiders, which possess a retrolateral tibial apophysis. Within the RTA clade, Agelena silvatica (Agelenidae) formed a sister clade to Oxytate striatipes (Thomisidae) which belongs to the Dionycha clade, characterized by the possession of two tarsal claws with tufts of hairs. The clade of the two species was sister to the Dionycha clade which includes Salticidae (Habronattus oregonensis and Telamonia vlijmi) and Selenopidae (Selenops bursarius). Uroctea compactilis (Oecobiidae), which belongs to the superfamily Eresoidea, formed a sister clade to the RTA clade.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim JY, Yoon JS, Park YC. 2016a. The complete mitochondrial genome of the green crab spider Oxytate striatipes (Araneae: Thomisidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1878–1879.
- Kim JY, Yoon KB, Park YC. 2016b. The complete mitochondrial genome of the jumping spider Telamonia vlijmi (Araneae: Salticidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:635–636.
- Kim YJ, Kim JP, Joo JH. 2003. Zoogeographic study of Korean spiders. Korean Arachnol. 19:207–225.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0b4a. Sunderland (MA): Sinauer Associates.
