Abstract
Schisandra sphenanthera (Austrobaileyales) is a famous traditional Chinese medicine being long-history used, is also one of early-diverging angiosperms and important links to uncover the early evolution of angiosperms. Here the complete mitochondrial genome of S. sphenanthera was obtained for the first time. It is 1,106,521 bp in length with 46.4% GC content. It contains 58 genes, including 41 protein coding genes, three ribosomal RNA genes and 14 transfer RNA genes. Phylogenetic analysis indicated that S. sphenanthera was placed in the basal angiosperm just after Amborella and Nuphar. The mitogenome of S. sphenanthera would provide a reliable genetic and evolutionary resource.
Schisandra sphenanthera Rehd. et E. H. Wilson, is a kind of liana and a horticultural plant with edible fruit in Austrobaileyales. Austrobaileyales is an early-diverging extant angiosperm branch just following Amborellales and Nymphaeales, thus provide an important link to uncover the origin and early evolution of angiosperms (APG IV 2016). Species in Schisandra are mainly native to east and southeast Asia but one species S. glabra in North America (Xia et al. Citation2009). Pharmacological studies on animals have shown that Schizandra affords a stress-protective effect against a broad spectrum of harmful factors (Iwata et al. Citation2004; Xiao et al. Citation2005; Panossian and Wikman Citation2008; Zhao et al. Citation2014; Jackson et al. Citation2017). Here we reported the complete mitochondrial genome (mitogenome) sequences of S. sphenanthera in Austrobaileyales for the first time. This would be helpful for the pharmaceutical, phylogenetic and mitochondrial evolutionary researches.
The whole genomic DNA was extracted from leaves of a mature S. sphenanthera plant which was growing in the natural forest habitat of Baoji city, Shaanxi province, China (34°05.313N107°42.315E). Voucher specimen and DNA sample (CHEN M. 20130709) were deposited in the herbarium of Institute of Botany, CAS (PE). Total 4.05 G 150 bp paired end raw reads using Illumina HiSeq2500 (Illumina, San Diego, CA) were generated and then trimmed low quality bases (<Q20) using NGS QC Toolkit (Patel and Jain Citation2012). Around 3.7 G trimmed reads were used to assemble mitogenome by Velvet v1.2.10 (Zerbino and Birney Citation2008). Contigs were connected manually in Geneious R8 (Biomatters, Inc., Auckland, New Zealand). Genome was annotated based on the former published mitogenomes of Liriodendron tulipifera (GenBank: NC_021152) and Nymphaea colorata (NC_037468). Plastid-derived DNAs were determined by searching against Schisandra chinensis chloroplast genome (NC_034908) with Blastn v2.4.0 (Camacho et al. Citation2009) and only fragments >200 bp were considered. Dispersed repeats were searched in Geneious with Repeat Finder v1.0 plugin, with settings minimum length 100 bp and maximum mismatch 5%.
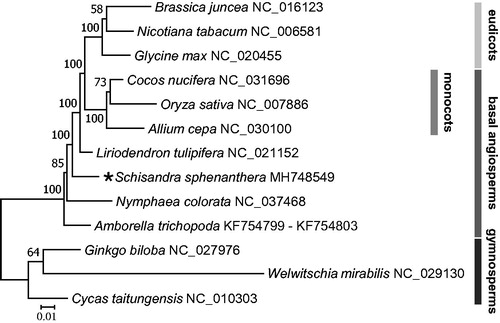
Figure 1. Phylogenetic tree based on 13 complete mitogenome sequences. The tree was constructed using RAxML method and bootstrap support value from 1000 replicates are shown above branches. All the mitogenome sequences are available in GenBank, the accession numbers are listed right to their scientific names.
The mitogenome of S. sphenanthera is 1,106,521 bp in length with 46.4% GC content and under the accession number MH748549. The mitogenome contains 58 genes, including 41 protein coding genes, three ribosomal RNA genes and 14 transfer RNA genes. Ten genes have introns and three of them are trans-spliced. Total plastid-derived DNA length is 26 kb, around 2.3% of the genome. Forty-six repeat pairs were found and nine of them are >500 bp. The total repeat length is around 74 kb.
A total of 41 mitochondrial coding genes of S. sphenanthera and another 12 seed plants were aligned with Mafft v7.306b (Yamada et al. Citation2016). The maximum likelihood tree was performed by using RAxML v8.2.10 (Stamatakis Citation2014) with 1000 bootstrap iterations in CIPRES website (Miller et al. Citation2010). The tree showed that S. sphenanthera places basal angiosperm branches just after Amborella (Amborellales) and Nuphar (Nymphaeales), which was consistent with the results by Angiosperm Phylogeny Group IV (APG IV 2016) (). The complete mitogenome of S. sphenanthera would represent a useful genetic resource to the further biological and evolutionary studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics. 10:421.
- Iwata H, Tezuka Y, Kadota S, Hiratsuka A, Watabe T. 2004. Identification and characterization of potent CYP3A4 inhibitors in Schisandra fruit extract. Drug Metab Dispos. 32:1351–1358.
- Jackson JP, Freeman KM, Friley WW, Herman AG, Black CB, Brouwer KR, Roe AL. 2017. Prediction of clinically relevant herb–drug clearance interactions using sandwich-cultured human hepatocytes: Schisandra spp. case study. Drug Metab Dispos. 45:1019–1026.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans 1–8.
- Panossian A, Wikman G. 2008. Pharmacology of Schisandra chinensis Bail.: an overview of Russian research and uses in medicine. J Ethnopharmacol. 118:183–212.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- [APG IV] The Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Xia NH, Liu YH, Saunders RMK. 2009. Schisandraceae. In: Wu ZY, Raven PH, editors. Flora of China, Vol. 7. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 39–47.
- Xiao WL, Zhu HJ, Shen YH, Li RT, Li SH, Sun HD, Zheng YT, Wang RR, Lu Y, Wang C, et al. 2005. Lancifodilactone G: a unique nortriterpenoid isolated from Schisandra lancifolia and its anti-HIV activity. Org Lett. 7:2145–2148.
- Yamada KD, Tomii K, Katoh K. 2016. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics. 32:3246–3251.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao T, Mao G, Zhang M, Feng W, Mao R, Zhu Y, Gu X, Li Q, Yang L, Wu X. 2014. Structure analysis of a bioactive heteropolysaccharide from Schisandra chinensis (Turcz.) Baill. Carbohydr Polym. 103:488–495.
