Abstract
Carnivorous plants have the ability to capture and digest small animals as a source of additional nutrients, which allows them to grow in nutrient-poor habitats. This study reports the complete mitochondrial genome sequence of a pitcher plant Nepenthes x ventrata. It was 520,764 bp in size with a GC content of 44.17% and contained 37 protein-coding genes, 2 pseudogenes, 18 tRNA genes and 3 rRNA genes. Four tRNA genes and the rps11 gene were probably transferred to mitochondrion form the chloroplast genome. Phylogenetic analysis confirmed that N. x ventrata belongs to the order Caryophyllales.
Carnivorous plants are able to attract, catch and digest their preys, usually insects, and assimilate nutrients for the growth (Adamec Citation1997). The obtained nutrients are primary used as source of nitrogen enabling these plants to survive in oligotrophic environments. Previous studies of organelle genomes of carnivorous plants of the genus Utricullaria (Lentibulariaceae) revealed the loss and pseudogenization of the NAD(P)H dehydrogenase genes in the chloroplast genome (Silva et al. Citation2016) while the structure and gene content of the mitochondrial genome remained typical for flowering plants (Silva et al. Citation2017).
We determined complete sequence of the mitochondrial genome (mtDNA) of the carnivorous plant Nepenthes x ventrata (Caryophyllales: Nepenthaceae), a natural hybrid between N. alata and N. ventricosa (Fleming Citation1979). Nepenthes use passive pitcher-shaped traps to catch small insects and are probably the most studied carnivorous plants (Mithöfer, Citation2011). Pitchers of Nepenthes were analysed by RNA-seq and proteomics for discovery of genetic trains related to carnivory (Hatano and Hamada Citation2008; Wan Zakaria et al. Citation2016).
N. x ventrata plants were grown in the greenhouse of Research Center of Biotechnology RAS, Moscow, Russia (55°41'58.4″N, 37°34'54.9″E). Total genomic DNA was extracted from leaves of a single plant (accession number NEP-CB1) and sequenced using a combination of Illumina and Nanopore techniques. The sequencing of a TrueSeq DNA library on Illumina HiSeq2500 generated 2.2 Gb high-quality cleaned sequences. 1,073,754 reads with a total length of 5 Gb were obtained on MinIon (Oxford Nanopore, UK). MinIon reads were de novo assembled using Miniasm 0.2 (Li Citation2016). Raw contigs were corrected by mapping MinIon reads back to the contigs using Minimap 0.2 and calling consensus with Racon 1.3.1 (Vaser et al. Citation2017). The final polishing was done with Illumina reads mapping using Pilon 1.22 (Walker et al. Citation2014). A single contig representing mtDNA was identified based on the comparison to other plant mitogenome sequences. mtDNA annotation was performed using MITOFY (Alverson et al. Citation2010) with further manual correction.
The mitochondrial genome of N. x ventrata (GenBank accession number MH798871) is a circular DNA molecule with 520,764 bp in length and a GC content of 44.17%. The genome contains 37 protein-coding genes, including ones for NADH dehydrogenase (nad1, 2, 3, 4, 4L, 5, 6, 7, 9), cytochrome c oxidase (cox1, 2, 3), cytochrome c biogenesis (ccmB, C, FN, FC), succinate dehydrogenase (sdh3, 4), apocytochrome b (cob), ATP synthase (atp1, 4, 6, 8, 9), ribosomal proteins (rpl2, 5 and rps1, 3, 4, 7, 10, 11, 12, 13, 19), maturase and membrane transporter MttB. 9 genes contained introns and 4 of them (rps3, nad1, nad2, nad5) were trans-spliced. In addition, the genome contains 18 tRNA genes coding for 15 amino acid, three rRNA genes (rrn5,18,26), and two pseudogenes (rpl16 and rps14). Four tRNA genes and the rps11 gene were likely acquired from the chloroplast genome; altogether plastid-like sequences accounted for ∼28 kb.
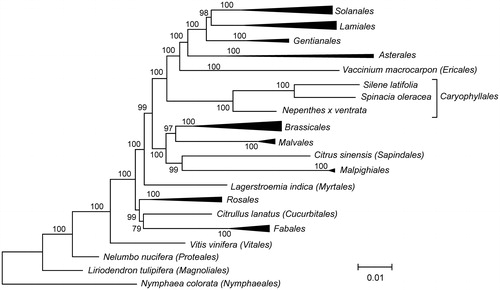
The maximum likelihood phylogenetic tree was based on concatenated sequences of exons of mitochondrial genes from N. x ventrata and other 42 species (). As expected, N. x ventrata is phylogenetically related to other members of the order Caryophyllales, Silene latifolia and Spinacia oleracea.
Figure 1. The maximum likelihood phylogenetic tree of Nepenthes x ventrata and 42 other angiosperm species. The tree is based on concatenated nucleotide sequences of exons of genes atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, cob, cox1, cox2, cox3, matR, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9, aligned using MAFFT v7.271 (Katoh et al., Citation2002). PhyML 3.1 (Guindon et al., Citation2010) was used to build the tree. Bootstrap support values are displayed on each node.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Adamec L. 1997. Mineral nutrition of carnivorous plants: a review. Bot Rev. 63:273–299.
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436–1448.
- Fleming R. 1979. Hybrid Nepenthes. Carniv Pl Newslett. 8:10–12.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Hatano N, Hamada T. 2008. Proteome analysis of pitcher fluid of the carnivorous plant Nepenthes alata. J Proteome Res. 7:809–816.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 32:2103–2110.
- Mithöfer A. 2011. Carnivorous pitcher plants: insights in an old topic. Phytochemistry. 72:1678–1682.
- Silva SR, Diaz YC, Penha HA, Pinheiro DG, Fernandes CC, Miranda VF, Michael TP, Varani AM. 2016. The chloroplast genome of Utricularia reniformis sheds light on the evolution of the ndh gene complex of terrestrial carnivorous plants from the Lentibulariaceae family. PLoS One. 11:e0165176.
- Silva SR, Alvarenga DO, Aranguren Y, Penha HA, Fernandes CC, Pinheiro DG, Oliveira MT, Michael TP, Miranda VFO, Varani AM. 2017. The mitochondrial genome of the terrestrial carnivorous plant Utricularia reniformis (Lentibulariaceae): structure, comparative analysis and evolutionary landmarks. PLoS One. 12:e0180484.
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27:737–746.
- Wan Zakaria WN, Loke KK, Goh HH, Mohd Noor N. 2016. RNA-seq analysis for plant carnivory gene discovery in Nepenthes × ventrata. Genom Data. 7:18–19.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9:e112963.
