Abstract
The characteristic of complete chloroplast (cp) genome of Magnolia sieboldii, an endangered species in China, was first sequenced in this study. It showed a typical quadripartite structure with a length of 160,177 bp. It composed of a large single copy (LSC) region of 88,238 bp, a small single copy (SSC) region of 18,763 bp, and a pair of inverted repeat (IR) regions of 26,588 bp. Further maximum parsimony phylogenetic analysis was performed based on 38 complete plastomes from 30 magnoliids and basal angiosperm species. The result supported a close relationship among M. grandiflora, M. officinals, M. sieboldii, and M. tripetala.
Magnolia sieboldii K. Koch is available most in the northern Magnolia species in China, it is distributed in the narrow geography regions and scattered in North China, Japan and North Korea (http://foc.eflora.cn/). It is an attractive horticultural and important economic tree species. It is already listed as a vulnerable species in China Species Red List and protected by the Chinese government as the third kind of rare and endangered plants as well. Here, we determined the complete chloroplast genome sequence of M. seiboldii.
Fresh young leaves of M. seiboldii were harvested from Tiannv mountain of Fushun County, Liaoning Province, China (41.5°N, 124.3°E). Six grams silica-gel dried young leaves were used for DNA extraction (Doyle and Dickson Citation1987). The complete cpDNA was sequenced following the method of Yang et al. (Citation2014). The genome of M. sieboldii was assembled using BioEdit software. Dual Organellar Genome Annotator (DOGMA) software was used to annotate the gene type of M. sieboldii (Wyman et al. Citation2004). The voucher specimens (Voucher number: HITBC-BRG-SY35347) of M. sieboldii were deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden (HITBC), Chinese Academy of Sciences.
The plastome of M. sieboldii (MH544144) shows a typical quadripartite structure with a length of 160,177 bp. It composes of a large single-copy (LSC) region of 88,238 bp, a small single copy (SSC) region of 18,763 bp, and a pair of inverted repeat (IR) regions of 26,588 bp. The GC-content of the whole plastome is 39.2%, and in LSC, SSC, and IRs regions are 37.9%, 34.3%, and 43.2%, respectively. The plastome contains a total of 113 genes, including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes.
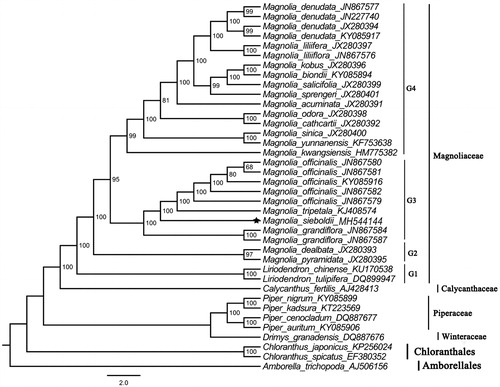
To determine the phylogenetic location of M. sieboldii with respect to the other 29 magnoliids and basal angiosperm species with fully sequenced chloroplast genomes, the complete plastome of M. sieboldii was used to reconstruct the phylogenetic relationships. Take the plastome of Amborella trichopoda (Amborellaceae, Amborellales; AJ506156) as an out-group, a maximum likelihood analysis based on the GTR + I + G model was performed with RAxML version 8 program using 1000 bootstrap replicates (Darriba et al. Citation2012; Stamatakis Citation2014). The phylogenetic tree in revealed that M. sieboldii was most closely related to M. grandiflora, M. officinals, and M. tripetala, which was in agreement with previous reports on the relationships among them (Azuma et al. Citation2001; Nie et al. Citation2008). Magnoliaceae was strongly supported as monophyletic (ML-BS = 100%), and four well-supported groups were recovered within these Magnolia species. In the Magnoliaceae, the basal group (ML-BS = 100%) including the two species of Liriodendron, the second group (ML-BS = 100%) including M. pyramidata and M.dealbata, the third group (ML-BS = 95%) including M. grandiflora, M. sieboldii, M. tripetala, and M. officinalis, and the core group (ML-BS = 95%) including M. kwangsiensis, M. yunnanensis, M. sinica, M. cathcartii, M. odora, M. acuminata, M. sprengeri, M. salicifolia, M. biondii, M. kobus, M. liliiflora, M. liliifera, and M. denudata.
Data availability
The plastome data of the M. sieboldii was submitted to Genebank of NCBI. The accession number from Genebank is MH544144.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Azuma H, García-Franco JG, Rico-Gray V, Thien LB. 2001. Molecular phylogeny of the Magnoliaceae: the biogeography of tropical and temperate disjunctions. Am J Botany. 88: 2275–2285.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 9:772.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36:715–722.
- Nie ZL, Wen J, Azuma H, Qiu YL, Sun H, Meng Y, Sun WB, Zimmer EA. 2008. Phylogenetic and biogeographic complexity of Magnoliaceae in the Northern Hemisphere inferred from three nuclear data sets. Mol Phylogenet Evol. 48:1027–1040.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.