Abstract
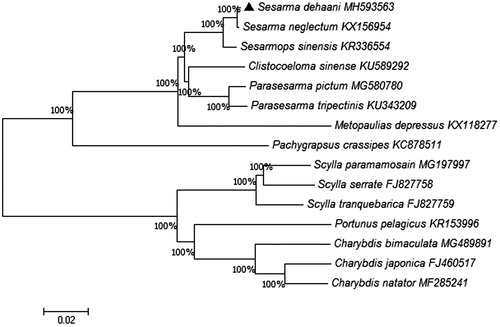
In this study, we first determined the complete mitochondrial genome of Sesarma dehaani. The mitogenome genome is a circular molecule of 15,917 bp, composed of 13 protein coding genes (PCGs), 22 tRNAs, two rRNAs, and one non-coding region. The heavy strand was predicted to have 9 PCGs, 14 tRNA genes, and a D-loop region, while the light strand was predicted to include 4 PCGs, 2 rRNA genes, 8 tRNA genes. ATG and TAA are the dominant initiation codon and stop codon, respectively. The phylogenetic analysis shows that S. dehaani and Sesarma neglectum have close relationships.
The estuarine crab, Sesarma dehaani, is the dominant species of invertebrate in the Sesarmidae family endemic to East Asia (Yang et al. Citation2014). It is called ‘ecosystem engineer’ because of its significant role in the ecological environment (Lee Citation1998). However, the population of S. dehaani has been declining, due to the large use of heavy metals in industrial and agricultural activities (Wang et al. Citation2014; Liu et al. Citation2015). Mitochondrial genomes (mitogenomes) play important roles in phylogenetic relationships analysis (Allcock et al. Citation2011).
The samples of S. dehaani (accession number B0040-1) were collected from Chongming, China (122°0′13.68″, 31°28′18.53). The specimens now were stored in East China Sea Environmental Monitoring Center (Shanghai, China). The genomic DNA isolated from the worker was sequenced. All the sequences were analyzed and assembled by BioEdit.
The complete mitogenome of S. dehaani (Genbank accession MH593563) was determined to be 15,917 bp long sequence, with the following nucleotide composition: A (37.46%), T (38.23%), C (14.83%), and G (9.49%). There are 8 overlapping regions, ranging from 1 to 7 bp. And 17 intergenic regions exist in this genome, ranging from 1 to 211bp. The gene content is similar to other crabs mitochondrial genomes (Park et al. Citation2017; Yang et al. Citation2017), comprising 13 protein coding genes (PCGs), 22 tRNAs, two rRNAs, and one non-coding region. Four PCGs (ND1, ND4, ND4L, and ND5), two rRNA genes (12S rRNA and 16S rRNA), and eight tRNA genes (His, Phe, Pro, Leu, Val, Gln, Cys, and Tyr) are encoded by L strand, while nine PCGs (ATP6, ATP8, ND2, ND3, ND6, COX1, COX2, COX3, and CYTB), fourteen tRNA genes (Ala, Arg, Asn, Asp, Glu, Gly, Ile, Leu, Lys, Met, two Ser, Thr, Trp), and a non-coding region (D-loop region) are encoded by H strand. Nine of thirteen PCGs start with the ATG typical initiation codon, three with ATT, and one with ATA. Eight PCGs use TAA and two PCGs use TAG as stop codon, whereas COX1, COX2, and CYTB use incomplete stop codon TA- or T–. The incomplete stop codon T– is commonly reported in some invertebrates (Masta and Boore Citation2004). There are 22 tRNAs genes in the mitogenome of S. dehaani, ranging from 63 to 73 bp. Non-coding region is A + T enrichment region, which is related to transcription and replication (Clayton Citation1992). The non-coding region of S. dehaani is located between 12S rRNA and tRNA-Gln genes, is 727 bp long. In addition, we detected two microsatellite-like (AT)8 elements in non-coding region, one at position of 13,663–13,679 bp, the other at position of 14,117–14,133 bp.
In order to analyze phylogenetic relationships, a phylogenetic tree was constructed based on 13 PCGs by neighbor-joining method (Kumar et al. Citation2016). Sesarma neglectum was grouped with S. dehaani as the sister species (). The S. dehaani mitogenome data provides additional genetic information for genetic analyses in the future study.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Allcock AL, Cooke IR, Strugnell JM. 2011. What can the mitochondrial genome reveal about higher‐evel phylogeny of the molluscan class Cephalopoda? Zool j Linnc Soc. 161:573–586.
- Clayton DA. 1992. Transcription and replication of animal mitochondrial DNAs. Int Rev Cytol. 141:217–232.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee SY. 1998. Ecological role of grapsid crabs in mangrove ecosystems: a review. Mar Freshwater Res. 49:335–343.
- Liu Z, Pan S, Sun Z, Ma R, Chen L, Wang Y, Wang S. 2015. Heavy metal spatial variability and historical changes in the Yangtze River estuary and North Jiangsu tidal flat. Mar Pollut Bull. 98:115–129.
- Masta SE, Boore JL. 2004. The complete mitochondrial genome sequence of the spider Habronattus oregonensis reveals rearranged and extremely truncated tRNAs. Mol Biol Evol. 21:893–902.
- Park YJ, Chang EP, Jung BK, Ibal JC, Jung YG, Hong SJ, Park GS, Lee SH, Ko HS, Shin JH. 2017. The first complete mitochondrial genome sequence of the leucosiid crab Pyrhila pisum (Arthropoda, Decapoda, Leucosiidae). Mitochondrial DNA Part B. 2:885–886.
- Wang J, Liu R, Zhang P, Yu W, Shen Z, Feng C. 2014. Spatial variation, environmental assessment and source identification of heavy metals in sediments of the Yangtze River Estuary. Mar Pollut Bull. 87:364–373.
- Yang CC, Møller AP, Ma ZJ, Li F, Liang W. 2014. Intensive nest predation by crabs produces source–sink dynamics in hosts and parasites. J Ornithol. 155:219–223.
- Yang X, Ma H, Waiho K, Fazhan H, Wang S, Wu Q, Shi X, You C, Lu J. 2017. The complete mitochondrial genome of the swimming crab Charybdis natator (Herbst) (Decapoda: Brachyura: Portunidae) and its phylogeny. Mitochondr DNA B. 2:530–531.