Abstract
In this study, we first sequenced and described the complete mitochondrial genome and phylogeny of Indicator xanthonotus. The whole genome of I. xanthonotus was 17,603 bp in length and contained 14 protein-coding genes, 22 transfer RNA genes, two ribosome RNA genes, and two non-coding control regions. The overall base composition of the mitochondrial DNA was 31.09% for A, 27.34% for T, 28.96% for C, and 12.61% for G, with a GC content of 41.57%. A phylogenetic tree strongly supported that I. xanthonotus clustered with another Piciformes species by high probability.
Keywords:
The Yellow-rumped Honeyguide (I. xanthonotus Blyth, 1842) is distributed in southern and southeastern of Asia (China, Nepal, Myanmar, et al.) and inhabits in rocky gorges and valleys with broadleaved or coniferous forest between 1450 and 3500 m (BirdLife International Citation2018; Gill and Donsker Citation2018). This species is a brood parasite avian and was usually considered to have strong association with Giant honey bee Apis dorsata (Fleischer Citation2011; BirdLife International Citation2018). This species is listed as Near Threatened (NT) on the IUCN Red List (IUCN, Citation2018). Webb and Moore (Citation2005) constructed a phylogenetic tree that analyzed intergeneric relationships combined 12S, Cyt b, and COI sequences. I. xanthonotus, as a scarce and poorly known species, very few studies had been reported, including complete mitochondrial genome not been revealed. Therefore, we sequenced the complete mitochondrial genome of I. xanthonotus to enhance our understanding on the phylogeny of Indicatoridae.
The specimen was collected from Mt. Gaoligong in Lushui County, which was located northwestern of Yunnan Province in China, and stored at College of Biodiversity Conservation and Utilization, Southwest Forestry University. The total mitochondrial DNA was extracted from the muscle tissue and sequenced using next-generation sequencing. The complete mitochondrial genome of I. xanthonotus was submitted to the NCBI database under the accession number MH737741. Phylogenetic tree of the relationships among Piciformes and Coraciiformes were presented using 16 species by maximum parsimony analyses using PAUP* version 4.0b10 software with 1000 bootstrap replication (Swofford Citation2002). Bayesian inference was calculated with MrBayes3.1.2 with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck Citation2003). Sequences of Coraciiformes (Eurystomus orientalis) obtained from GenBank (NC_011716.2) were used as outgroups to root trees following Mccormack et al. (Citation2013) and Eo (Citation2017).
The complete mitochondrial genome of I. xanthonotus was 17,603 bp in length. A total of 40 mitochondrial genes were identified, including 14 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and two non-coding control regions (D-loop). Among these genes, ND6 and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu) were located on the light strand (Lstrand), while all of the remaining genes were located on the heavy strand (H-strand). The overall base composition of A. insignis mitogenome was 31.09% for A, 12.61% for G 28.96% for C, and 27.34% for T, A + T content is 58.43%, which is higher than G + C content of 41.57%, similar to other Piciformes (Fuchs et al. Citation2015; Zhang et al. Citation2016; Eo Citation2017).
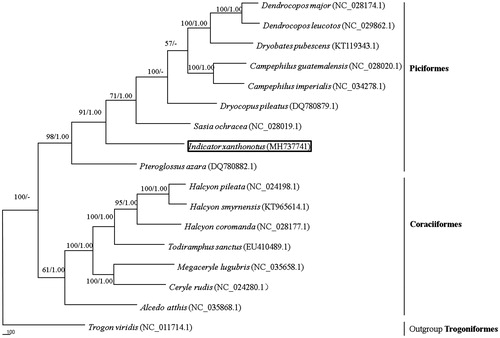
The reconstructed phylogenetic tree supported the placement of I. xanthonotus in Piciformes (). In our results, 16 species were clustered into two groups. Indicator xanthonotus clustered with other Piciformes species and was strongly supported by the analyses of protein-coding genes. Thus, the mitochondrial genome reported here would be useful in the current understanding of the phylogeny and evolution of Piciformes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- BirdLife International. 2018. Species factsheet: Indicator xanthonotus. [accessed 2018 Aug 14]. http://www.birdlife.org.
- Eo SH. 2017. Complete mitochondrial genome of white-backed woodpecker Dendrocopos leucotos (piciformes: picidae) and its phylogenetic position. Mitochondrial DNA Part B. 2:451–452.
- Fleischer RC. 2011. Ladies and gentes: maternally inherited DNA and ancient honeyguide host races. Proc Natl Acad Sci USA. 108:17859–17860.
- Fuchs J, Pons JM, Pasquet E, Bonillo C. 2015. Complete mitochondrial genomes of the white-browed piculet (Sasia ochracea, picidae) and pale-billed woodpecker (Campephilus guatemalensis, picidae. Mitochondrial DNA. 27:1–2.
- IUCN. 2018. The IUCN Red List of Threatened Species. Version 2018-1. [accessed 2018 Aug 14]. www.incnredlist.org
- Mccormack JE, Harvey MG, Faircloth BC, Crawford NG, Glenn TC, Brumfield RT. 2013. A phylogeny of birds based on over 1,500 loci collected by target enrichment and high-throughput sequencing. Plos One. 8:e54848.
- Gill F, Donsker D. 2018. IOC World Bird List (v8.2). [accessed 2018 August 14]. http://www.worldbirdnames.org/
- Ronquist F, Huelsenbeck JP. 2003. MrBayes3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Swofford DL. 2002 PAUP*: Phylogenetic analysis using parsimony (*and other methods). Version4.0b10. Sinauer Associates, Massachusetts.
- Webb DM, Moore WS. 2005. A phylogenetic analysis of woodpeckers and their allies using 12S, Cyt b, and COI nucleotide sequences (class aves; order piciformes). Mol Phylogenet Evol. 36:233–248.
- Zhang Z, An M, Deng Y, Zhu S. 2016. The complete mitochondrial genome of the Downy Woodpecker Picoides pubescens (Piciformes: Picidae). Mitochondrial DNA Part A. 27:3479–3480.