Abstract
Pseudostellaria longipedicellata which belongs to Caryophyllaceae is endemic in South Korea. In this study, we presented first complete chloroplast genome of P. longipedicellata which is 149,626 bp and has four sub-regions: 81,292 bp of large single copy (LSC) and 16,984 bp of small single copy (SSC) regions are separated by 25,675 bp of inverted repeat (IRs) regions including 126 genes (81 CDS, eight rRNAs, and 37 tRNAs). The overall GC contents of the chloroplast genome were 36.5% and in the LSC, SSC and IR regions were 34.3%, 29.3%, and 42.4%, respectively.
Pseudostellaria Pax (Caryophyllaceae) is a small genus distributed in temperate region. It consists of 25 species presenting high diversity in Asia. Two of three species distributed in North America have been separated into a new genus Hartmaniella M.L.Zhang & Rabeler but phylogenetic position of the other species P. jamesiana is still unclear forming a clade with Stellaria americana. Except these three species, core Pseudostellaria divided into two clades is monophyletic (Zhang et al. Citation2017).
Pseudostellaria longipedicellata S. Lee, K. Heo & S. C. Kim was first announced as new species in 2012(Lee et al. Citation2012). Morphological characters of P. longipedicellata are closely related to those of Psedusotellaria palibiniana and Psedusotellaria okmotoi. These are distinguished from P. longipedicellata by shorter pedicel and puberulent pedicels, respectively (Ohwi Citation1937; Choi Citation1999) and by being distributed allopatically between P. longipedicellata and the rest of species (unpublished data). Here, we presented the complete chloroplast genome sequence of P. longipedicellata as a first chloroplast genome in Pseudostellaria genus.
Pseudostellaria longipedicellata was collected in Mt. Taebaek, Gangwon Province, South Korea (Voucher in Cyber Herbarium at InfoBoss; Y. Kim, IB-00654). Total DNA was extracted from fresh leaves of P. longipedicellata using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq 4000 (Macrogen Inc, Seoul, Korea), and de novo assembly was done using Velvet 1.2.21 (Zerbino and Birney Citation2008). Geneious R8 v8.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation.
The chloroplast genome of P. longipedicellata (MH373593) is 149,626 bp and has four sub-regions: 81,292 bp of large single copy (LSC) and 16,984 bp of small single copy (SSC) regions are separated by 25,765 bp of inverted repeat (IRs). It contained 126 genes (81 CDS, 8 rRNAs, and 37 tRNAs); 18 genes (seven CDS, four rRNAs, and seven tRNAs) are duplicated in inverted repeat regions. The overall GC content of P. longipedicellata is 36.5% and in the LSC, SSC, and IR regions were 34.3%, 29.3%, and 42.4%, respectively.
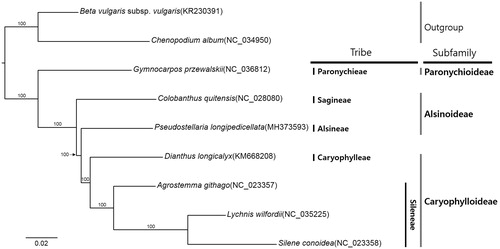
Seven complete chloroplast genomes of Caryophyllaceae and two species from Amaranthaceae were used for constructing maximum likelihood method phylogenetic tree (Alignment was conducted using MAFFT v7.388 (Katoh and Standley Citation2013) and phylogenetic tree was constructed using IQ-TREE v1.6.6 (Nguyen et al. Citation2014)). The tree shows that genus Pseudostellaria is sister to Caryophylloideae, forming paraphyletic clade in Alsinoideae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Choi K. 1999. Biosystematic Study of Pseudostellaria Pax (Caryophyllaceae) [doctoral thesis]. Gyeongbuk National University.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lee S, Heo K-I, Kim S-C. 2012. A new species of Pseudostellaria (Caryophyllaceae) from Korea. Novon. 22:25–31.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ohwi J. 1937. The revision of the genus Pseudostellaria. Jap J Bot. 9:95–105.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang M-L, Zeng X-Q, Li C, Sanderson SC, Byalt VV, Lei Y. 2017. Molecular phylogenetic analysis and character evolution in Pseudostellaria (Caryophyllaceae) and description of a new genus, Hartmaniella, in North America. Bot J Linnean Soc. 184:444–456.