Abstract
The first complete chloroplast genome sequences of Korean endemic pokeweed in Ulleung Island, Phytolacca insularis, were reported in this study. The P. insularis plastome was 156,419 bp long, with the large single copy (LSC) region of 86,106 bp, the small single copy (SSC) region of 18,335 bp, and two inverted repeat (IR) regions of 25,989 bp. The plastome contained 132 genes, including 84 protein-coding, eight ribosomal RNA, and 39 transfer RNA genes. The overall GC content was 36.8%. Phylogenetic analysis of 13 representative plastomes within the order Caryophyllales suggests that P. insularis is closely related to the species in family Aizoaceae.
The genus Phytolacca L. belongs to family Phytolaccaceae and consists of about 25 species of perennial herbs, shrubs and rarely trees. The genus shows nearly cosmopolitan distribution, mostly native to South America and a few species in Africa and Asia (Nowicke Citation1969; Shu Citation2003). Phytolacca is a pharmaceutically important genus owing to several active compounds in leaves and fruits for analgesic, anti-inflammatory, bactericidal, and fungicidal actions (Hernandez et al. Citation2013). In particular, P. acinosa Roxb. in East Asia is known as ‘Phytolaccae Radix’, one of the important oriental herbal medicines for diverse biological properties, such as antibacterial, anti-inflammatory, antidiuretic, antiviral, and anticancer (Zhang et al. Citation1990; Gao et al. Citation2009). Also, berries and young leaves were utilized as an adulterant of red wine and made into poke salad, respectively (Nowicke Citation1969). Of 25 species of Phytolacca, one native (P. insularis Nakai) and two introduced species (P. acinosa and P. americana L.) are known in Korea (Lee, Citation2004). Phytolacca insularis occurs exclusively on Ulleung Island in East Sea (Nakai Citation1918; Kim Citation2004). Taxonomic status as a distinct insular endemic and relationship to other congeneric species are highly controversial. For example, Hong (Citation2007) suggested that P. insularis is closely related to P. japonica Makino based on vegetative and reproductive traits, while Chae et al. (Citation2007) recognized P. insularis as a form of P. acinosa based on pollen and seed morphology and ITS sequences. Thus, it is yet to be determined the distinct taxonomic status of P. insularis and phylogenetic relationships within the genus. For the first time, we sequenced the complete plastome of Phytolacca, P. insularis, and assessed phylogenetic position within Caryophyllales.
Total DNA (Voucher specimen: N37°30′26.4″ E130°49′24.7″, Lee171010135, KNU) was isolated using the DNeasy plant Mini Kit (Quiagen, Carlsbad, CA) and sequenced by the Illumina HiSeq 4000 (Illumina Inc., San Diego, CA). A total of 3,664,7851 paired-end reads were obtained and assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008). The tRNAs were confirmed using tRNAsacn-SE (Lowe and Eddy Citation1997). The total plastome length of P. insularis (MH376309) was 156,419 bp, with large single copy (LSC; 86,106 bp), small single copy (SSC; 18,335 bp), and two inverted repeats (IRa and IRb; 25,989 bp each). The overall GC content was 36.8% (LSC, 34.7%; SSC, 30.4%; IRs, 42.7%) and the plastome contained 132 genes, including 84 protein-coding, eight rRNA, and 39 tRNA genes. A total of 19 genes were duplicated in the inverted repeat regions including eight tRNA, four rRNA, and six protein coding genes. The complete ycf1 gene was included in the IR at the SSC/IRa junction and the complete infA gene was located in LSC. The partial ycf1 gene became a pesudogene and located at IRb/SSC junction.
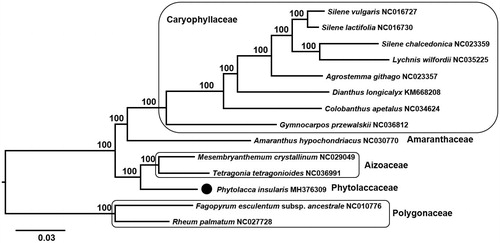
To confirm the phylogenetic position of P. insularis, 13 representative species of Caryophyllales were aligned using MAFFT v.7 (Katoh and Standley Citation2013) and maximum likelihood (ML) analysis was conducted using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015). The ML tree showed that P. insularis (Phytolaccaceae) is sister to Aizoaceae ().
Additional information
Funding
References
- Chae SH, So SK, Han KS, Kim MY, Park SH, Lee JK. 2007. A taxonomic review of Phytolacca insularis (Phytolaccaceae). Korean J Pl Taxon. 37:431–446.
- Gao HM, Liu JX, Wang ZM, Wang WH. 2009. Phytolacacinoside A, a new triterpenoid saponin from Phytolacca acinosa Roxb. J Asian Nat Prod Res. 11:433–438.
- Hernández M, Murae M, Ringuelet J, Petri I, Gallo D, Arambarri A. 2013. Effect of aqueous and alcohol extracts of Phytolacca tetramera (Phytolaccaceae) leaves on Colletotrichum gloeosporioides (Ascomycota). Bol Soc Argent Bot. 48:201–209.
- Hong SP. 2007. Phytolaccaceae. In: Flora of Korea Editional Commitee (eds), The genera of vascular plants of Korea. Seoul: Academy Publishing Co., pp. 285–286.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim MY. 2004. Korean endemic plants. Seoul: Solbook Publishing Co., pp. 72–73.
- Lee TB. 2004. Coloured flora of Korea. Seoul: KyoHak Publishing Co., pp. 51.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Nakai T. 1918. Phytolacca insularis Nakai. Bot Mag Tokyo. 32:217.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Nowicke JW. 1969. Palynotaxonomic study of the Phytolaccaceae. Ann Miss Bot Gard. 55:94–363.
- Shu SL. 2003. Phytolacca. In: Wu ZY, Raven PH and Hong DY (eds), Flora of China. Vol. 5. St Louis: Science Press, Beijing and Missouri Botanical Garden Press, pp. 435–436.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang J, Qian D, Zheng Q. 1990. Effects of Phytolacca acinosa polysaccharides I on cytotoxicity of macrophages and its production of tumor necrosis factor and interleukin 1. Chin J Cancer Res. 2:13–16.