Abstract
Bixa orellana is a small tree known for its red, oil-soluble pigment contained in the seed coat that is used as a natural dye and food coloring. In this study, we assembled and characterized the complete chloroplast genome of B. orellana as a resource for future genetic studies. With a total length of 159,825 bp, the chloroplast genome comprised of a large single-copy (LSC) region of 89,476 bp, a small single-copy (SSC) region of 19,617 bp, and two inverted repeat (IR) regions of 25,356 bp each. A total of 127 genes were predicted, consisting of 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis confirmed the position of B. orellana within the order Malvales.
Bixa orellana Linnaeus is an evergreen shrub or small tree known for its red, oil-soluble pigment (bixin) that is contained in the seed coat. As one of the five-member species of the genus, it is native to tropical America but widely cultivated in tropical regions of the world (eFloras Citation2008). Aside from the red pigment from the seed that is usually used as natural dyes and food coloring, the different parts of the plant are also used as traditional medicine in many cultures (de Araujo Vilar et al. Citation2014). As the best-known species in the family Bixaceae, in this study, we characterized the complete chloroplast genome sequence of B. orellana as a resource for future genetic studies on this and other related species.
Leaf samples of B. orellana were obtained from the Guangzhou Institute of Forestry and Landscape Architecture, with voucher specimen also deposited at the Sun Yat-sen University Herbarium (SYS; specimen code SYS-Bore-2018-01). After DNA extraction, a library with the insertion size of ∼400 bp was constructed, and high-throughput DNA sequencing (pair-end 150 bp) was performed on an Illumina Hiseq 2500 platform, generating approximately 4 Gb of sequence data. Using a B. orellana partial rbcL gene sequence (Y15139.1) as seed and the Gossypium arboretum chloroplast genome sequence (HQ325740.1) as reference, the chloroplast genome was assembled from the Illumina data using the program NOVOPlasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome sequence was then annotated using DOGMA (Wyman et al. Citation2004) and manually corrected.
The complete chloroplast genome sequence of B. orellana (GenBank accession MH751592) was 159,825 bp in length, with a large single-copy (LSC) region of 89,476 bp, a small single-copy (SSC) region of 19,617 bp, and two inverted repeat (IR) regions of 25,356 bp each. A total of 127 genes were predicted, consisting of 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content was 36.40%.
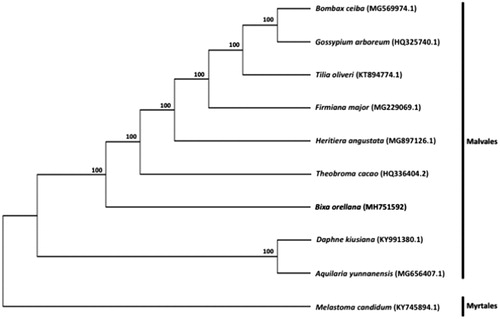
To understand the phylogenetic position of Bixa within the order Malvales, chloroplast genome sequences of eight other representative species in Malvales (Aquilaria yunnanensis, Bombax ceiba, Daphne kiusiana, Firmiana major, Gossypium arboretum, Heritiera angustata, Theobroma cacao, Tilia oliveri), as well as Melastoma candidum of the order Myrtales, were downloaded from the NCBI GenBank database for phylogenetic analysis. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with M. candidum as outgroup. As shown in , B. orellana clustered within Malvales with strong bootstrap support.
Figure 1. Maximum likelihood tree showing the relationship among Bixa orellana and representative species within the order Malvales, based on whole chloroplast genome sequences, with Melastoma candidum (Myrtales) as outgroup. Shown next to the nodes are bootstrap support values based on 1000 replicates.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- de Araujo Vilar D, de Araujo Vilar MS, de Lima e Moura TFA, Raffin FN, de Oliveira MR, de Oliveira Franco CF, de Athayde-Filho PF, de Fatima Formiga Melo Diniz M, Barbosa-Filho JM. 2014. Traditional uses, chemical constituents, and biological activities of Bixa orellana L.: a review. Sci World J. 2014:Article ID 857292.
- eFloras. 2008. Bixa orellana. Flora of China 13: 71. [accessed 2018 August 15]. http://www.efloras.org.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
