Abstract
In this study, we present the first complete mitochondrial genomes of two species of the genus Ovis – snow sheep (Ovis nivicola) and thinhorn sheep (Ovis dalli). The mitochondrial genomes have a total length of 16,471 and 16,464 bp, respectively, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding region. Phylogenetic analyses using nucleotide sequences of the 13 PCGs indicated that O. nivicola and O. dalli clustered with another representative of the Pachyceros subgenus – O. canadensis, which was consistent with the previous morphological classification.
The genus Ovis is divided into two subgenus – Ovis, which includes argali (O. ammon), urial (O. vignei), Asiatic mouflon (O. orientalis), European mouflon (O. orientalis musimon), Cyprus mouflon (O. orientalis ophion), and domestic sheep (O. aries), and Pachyceros, which includes snow sheep (O. nivicola), thinhorn (O. dalli), and bighorn sheep (O. canadensis). To date, complete mitogenomes of the Pachyceros sheep were represented only by two sequences of O. canadensis (Miller et al. Citation2012; Davenport et al. Citation2018). Here, we present the first complete mitochondrial genomes of O. nivicola and O. dalli. Both sequences were deposited in GenBank under accession numbers: MH779626 and MH779627, respectively.
For this study, a specimen of O. nivicola was collected from the Chersky Range, Russia (64°24′35″N; 143°17′08″E) and a specimen of O. dalli was collected from the Alaska Range, USA (63°48′6″N, 147°33′24″W). The samples were deposited in the biobank at the L.K. Ernst Federal Science Center for Animal Husbandry, Russia (Biobank State Registration No. 498808). Genomic DNA was extracted from muscle tissue using a Nexttec column (Nexttec Biotechnology GmbH, Germany). For next-generation sequencing of mtDNA, three primer pairs were placed in well-conserved regions and three overlapping (overlap > 290 bp) PCR products were produced. These products (6.5, 5.7, and 6.7 kb) were separately amplified, purified, and used for the preparation of sequencing libraries, which were then sequenced by 100-bp paired-end procedure on a HiSeq 1500 (Illumina). The hypervariable part of the control region was sequenced by Sanger technology using primers designed according to the flanking sequences assembled from the NGS data. Mitos WebServer (Bernt et al. Citation2013) was used to annotate the mitochondrial genomes. The number of tandem repeats was estimated with the Tandem Repeats Finder 4.09 (Benson Citation1999). The DNA sequences were aligned using the MUSCLE algorithm (Edgar Citation2004), as implemented in MEGA 7.0.26 (Kumar et al. Citation2016). PartitionFinder 2 (Lanfear et al. Citation2017) was used to select the best partitioning scheme and evolutionary models.
The whole mitochondrial genomes of O. nivicola and O. dalli are 16,471 bp and 16,464 bp in size, respectively. The gene arrangement pattern in both species is similar to the typical vertebrate mitochondrial genome, containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding region. We found two 75 bp tandem repeats in the control regions of the newly assembled mitogenomes, which shows that all species of the Pachyceros subgenus are characterized by the same number of tandem repeats.
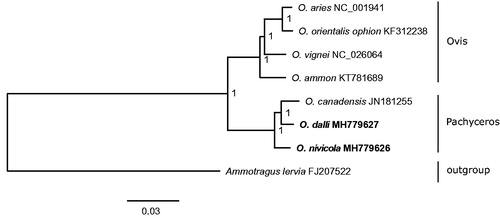
Phylogenetic analysis () using the Bayesian inference method in MrBayes 3.2.6 (Ronquist et al. Citation2012) was performed on the concatenated datasets of 13 PCGs and 2 rRNAs of the newly sequenced mitogenomes, five other Ovis species downloaded from GenBank (Hiendleder et al. Citation1998; Miller et al. Citation2012; Lv et al. Citation2015; Sanna et al. Citation2015; Gan Citation2016) and the barbary sheep (Ammotragus lervia) sequence (Hassanin et al. Citation2009) taken as an outgroup. It was shown that O. nivicola and O. dalli clustered in the Pachyceros clade. Our results are consistent with the previous morphological classification.
Figure 1. Bayesian tree based on the concatenated nucleotide sequences of 13 mitochondrial PCGs and two mRNA indicating evolutionary relationships of the genus Ovis. Node numbers show posterior probability values. GenBank accession numbers are given with species names.
The study was supported by the Russian Science Foundation (Project no.14–36–00039). The biobank collection (State Registration No. 498808) is supported by the Federal Agency for Scientific Organizations, Russia. We would like to thank Edward V. Bendersky, Viktor N. Kim and the Mountain Hunters Club (http://kgo-club.ru) for providing the sample of O. dalli.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Davenport KM, Duan M, Hunter SS, New DD, Fagnan MW, Highland MA, Murdoch BM. 2018. Complete mitochondrial genome sequence of bighorn sheep. Genome Announc. 6:e00464–e00418.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Gan SQ, Xu MS, Meng JM, Tang H, Li J, Yuan W, Wang X, Wang L, Zhou P. 2016. The complete mitochondrial genome of Marco Polo wild sheep (Ovis ammon polii). Mitochondrial DNA Part B. 1:92–93.
- Hassanin A, Ropiquet A, Couloux A, Cruaud C. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol. 68:293–310.
- Hiendleder S, Lewalski H, Wassmuth R, Janke A. 1998. The complete mitochondrial DNA sequence of the domestic sheep (Ovis aries) and comparison with the other major ovine haplotype. J Mol Evol. 47:441–448.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Lv FH, Peng WF, Yang J, Zhao YX, Li WR, Liu MJ, Ma YH, Zhao QJ, Yang GL, Wang F, et al. 2015. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol Biol Evol. 32:2515–2533.
- Miller JM, Malenfant RM, Moore SS, Coltman DW. 2012. Short reads, circular genome: skimming SOLiD sequence to construct the bighorn sheep mitochondrial genome. J Hered. 103:140–146.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systemat Biol. 61:539–542.
- Sanna D, Barbato M, Hadjisterkotis E, Cossu P, Decandia L, Trova S, Pirastru M, Leoni GG, Naitana S, Francalacci P, et al. 2015. The first mitogenome of the Cyprus mouflon (Ovis gmelini ophion): new insights into the phylogeny of the genus Ovis. PLoS One. 10:E0144257.
