Abstract
We obtained the complete mitogenome of Proschkinia sp. strain SZCZR1824, a strain belonging to a poorly known diatom genus with no previous molecular data. This genome is 48,863 bp long, with two group I introns in rnl and three group II introns in cox1. Using mitogenomic data, Proschkinia sp. was recovered with Fistulifera solaris, far distant from Navicula and Nitzschia, two genera with which Proschkinia has sometimes been associated based on morphology.
Proschkinia is a rare genus of diatom of controversial higher classification. Originally classified as a relative of Nitzschia on the basis of light microscopy (Karayeva Citation1978), Proschkinia is now classified within its own family, Proschkiniaceae (Round et al. Citation1990), within the Naviculales, without any specific hypothesis as to its relationship with other Naviculales. Additional SEM studies failed to further resolve you need the relationship of Proschkinia (Brogan and Rosowski Citation1988; Cox Citation1988, Citation2012).
We sequenced the complete mitochondrial genome of Proschkinia sp. strain SZCZR1824, a strain displaying similarities with Proschkinia complanatoides, and created a comparative data set from this and published diatom mitochondrial genes in order to better resolve the phylogenetic position of Proschkinia.
Proschkinia sp. strain SZCZR1824, originating from Padori Beach on the Yellow Sea coast of Korea (36°44′15.0″N, 126°07′49.7″E) was obtained from Kunsan National University (Korea). Total DNA was extracted following Doyle and Doyle (Citation1990). Paired-end sequencing (150 bp) was conducted by the Beijing Genomic Institute (Shenzhen) on HiSeq 4000, with inserts of 300 bp, for a total of ca. 30 million reads. Assembly was performed using Ray 2.3.1 (Boisvert et al. Citation2010) with a k-mer of 35. Gene identification was done using custom tools developed at Laval University (Gagnon Citation2004).
A permanent slide with cleaned frustules of SZCZR1824 is kept in the collection of the University of Szczecin. Frozen DNA and pellets of cells are also being stored in Szczecin at −20 °C.
The mitogenome of Proschkinia sp. SZCZR1824 (MH800316) is 48,863 bp long and encodes two rRNAs, 22 tRNAs and 33 proteins, for a total of 57 gene products. In addition, there are two free-standing open reading frames (orf143 and orf243) with no obvious function. The large subunit rRNA gene (rnl) is interrupted by two group I introns, whereas cox1 is interrupted by three large group II introns. Each of the cox1 introns contains a putative reverse transcriptase gene (orf714, orf789 and orf1002) and BlastP searches using these gene products as queries identified putative diatom proteins encoded by cox1 introns: YP_009495514 (Psammoneis japonica) for orf714, YP_009144752 (Pseudo-nitzschia multiseries) for orf789 and AVR57660 (Halamphora sp.) for orf1002.
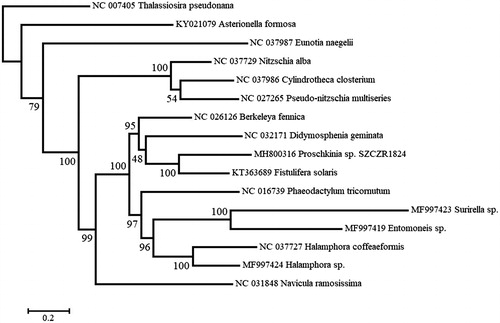
A maximum-likelihood phylogenetic analysis was performed on a concatenated data set of cox1, cox2, cox3, cob, nad2, nad4, nad5, and nad11 from 16 diatoms using RAxML 8.2.12 (Stamatakis Citation2014). Proschkinia sp. was recovered as sister to Fistulifera solaris (), which was previously placed in the Stauroneidaceae (Cox Citation2015) or Naviculaceae (NCBI taxonomy, accessed 25 August 2018) in the Naviculales. These two species formed a larger, strongly supported clade with Berkeleya fennica (Berkeleyales) and Didymosphenia geminata (Cymbellales). Bacillariales species (Nitzschia, Pseudo-nitzschia and Cylindrotheca) and Navicula ramosissima are separated by several nodes from Proschkinia, a result incongruent with the taxonomic placements reported for the latter diatom by Karayeva (Citation1978) and Round et al. (Citation1990). Fistulifera, like Proschkinia, possesses a special structure, called a ‘fistula’, between the raphe slots at the valve center (Lange-Bertalot Citation1997; Zgrundo et al. Citation2013). Based on this shared feature and the mitochondrial phylogeny presented here, we propose that Fistulifera and Proschkinia belong to the same family, Proschkiniaceae.
Figure 1. Maximum likelihood phylogeny obtained on concatenated mitochondrial genes (cox1, cox2, cox3, cob, nad2, nad4, nad5, and nad11) of Proschkinia sp. and other diatoms, with Thalassiosira pseudonana being the outgroup. Numbers next to nodes are support values obtained after 100 bootstrap replicates.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17:1519–1533.
- Brogan MW, Rosowski JR. 1988. Frustular morphology and taxonomic affinities of Navicula complanatoides (Bacillariophyceae). J Phycol. 24:262–273.
- Cox EJ. 1988. Taxonomic studies on the diatom genus Navicula V. The establishment of Parlibellus gen. nov. for some members of Navicula sect. Microstigmaticae. Diat Res. 3:9–38.
- Cox EJ. 2012. Ontogeny, homology, and terminology-wall morphogenesis as an aid to character recognition and character state definition for pennate diatom systematics. J Phycol. 48:1–31.
- Cox EJ. 2015. Diatoms, Diatomeae (Bacillariophyceae s.l., Bacillariophyta). In: Frey W, editor. Syllabus of plant families. A. Engler’s Syllabus der Pflanzenfamilien, 13th ed. Part 2/1: Photoautotrophic Eukaryotic Algae. Glaucocystophyta, Cryptophyta, Dinophyta/Dinozoa, Haptophyta, Heterokontophyta/Ochrophyta, Chlorarachniophyta/Cercozoa, Euglenophyta/Euglenozoa, Chlorophyta, Streptophyta p.p. Stuttgart: Borntraeger Science Publishers; p. 64–103.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Gagnon J. 2004. Création d'outils pour l'automatisation d'analyses phylogénétiques de génomes d'organites [Development of bioinformatic tools for the phylogenetic analyzes of organellar genomes] [M.Sc. dissertation]. University of Laval.
- Karayeva NI. 1978. Novyi podporyadok diatomovykh vodoroslei (a new suborder of diatoms). Botanischeskii Zhurnal. 63:1747–1750.
- Lange-Bertalot H. 1997. Frankophila, Mayamaea und Fistulifera: drei neue Gattungen der Klasse Bacillariophyceae. Archiv Protisten. 148:65–76.
- Round FE, Crawford RM, Mann DG. 1990. The diatoms Biology and morphology of the genera. Cambridge: Cambridge University Press; pp. [i–ix], 1–747.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zgrundo A, Lemke P, Pniewski F, Cox EJ, Latała A. 2013. Morphological and molecular phylogenetic studies on Fistulifera saprophila. Diat Res. 28:431–443.
