Abstract
The complete chloroplast genome from Elymus sibiricus Linn, an important forage grass of the Gramineae, is determined in this study. The whole chloroplast genome sequence of E. sibiricus Linn has been characterized by Illumina pair-end sequencing. The circular genome is 135,074 bp long, containing a large single copy region (LSC) of 80,681 bp and a small single copy region (SSC) of 12,767 bp, which are separated by a pair of 20,813 bp inverted repeat regions (IRs). It encodes a total of 142 genes, including 86 protein-coding genes (81 PCG species), 48 tRNA genes (30 tRNA species) and eight ribosomal RNA genes (four rRNA species). The most of gene species occur as a single copy, while 16 gene species occur in double copies. The overall A + T content of is 61.7%, while the corresponding values of the LSC, SSC and IR regions are 63.6%, 67.8% and 56.0%, respectively. Phylogenetic analysis suggested that E. sibiricus Linn was relatively close to Connorochloa tenuis compared with species of other generain Gramineae among the species analyzed. This complete chloroplast genome will provide valuable insight into conservation and exploitation efforts for this species as well as useful resources for studying the Gramineae phylogeny.
Elymus sibiricus Linn, is an endemic self-fertilizing species of the Gramineae mainly distributed in northern Asia (Nevski Citation1934).This plant has not only been widely utilized for forage grass in some temperate regions, especially in the Qinghai–Tibet plateau, China (Lei et al. Citation2014), but also has a well adaptation such as high disease resistance and cold, drought and salinity stress tolerance (Löve Citation1982; Motsny and Simonenko Citation1996; Lei et al. Citation2014). However, it is now threatened and experiencing a rapid population decline due to warming climate and excessive grazing (Lei et al. Citation2014). In addition, E. sibiricus Linn has special taxonomic significance as it is the type species of the genus Elymus distributed in most temperate regions of the world (Dewey Citation1984; Lei et al. Citation2014). Therefore, this species needs to be better studied and prioritized as a conservation and exploitation target. Analyzing the chloroplast genome proved to be an efficient approach to shed light on plant molecular systematics (Moore et al. Citation2010; Zhang et al. Citation2011). In the present study, the complete chloroplast genome sequence of E. sibiricus Linn will contribute effective use and provide useful resources for better understanding the evolution of the whole grass family (Gramineae).
Genomic DNA was extracted from fresh leaves of an individual of E. sibiricus Linn collected from the Qinghai–Tibet plateau (Qinghai Province, China; the specimen was deposited at Qinghai University; accession number: DCX-2018-QM1-3). The genomic library was sequenced on an Illumina Hiseq X Ten platform. 10 million high-quality reads were assembled into complete chloroplast genome by using Velvet (Zerbino and Birney Citation2008).The assembled chloroplast genome was annotated using Geneious (Kearse et al. Citation2012). Then, submitted to GenBank (accession no. MH732740). The chloroplast sequence of the E. sibiricus Linn was 135,074 bp, including the large single copy region (LSC, 80,681 bp), the small single copy region (SSC,12,767 bp), and a pair of 20,813 bp inverted repeat regions (IRs). The circular genome contained 142 genes, including 86 protein-coding genes (81 PCG species), eight ribosomal RNA genes (four rRNA species) and 48 tRNA genes (30 tRNA species). The most of gene species occurred in a single copy, while 16 gene species occurred in double copies, including four rRNA species (4.5S, 5S, 16S and 23S rRNA), seven tRNA species and five PCG species. The overall A + T content of the circular genome was 61.7%, while the corresponding values of the LSC, SSC and IR regions were 63.6, 67.8 and 56.0%, respectively.
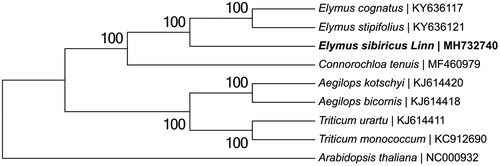
A neighbor-joining phylogenetic tree was constructed based on the concatenated coding sequences of chloroplast genomes for a panel of nine species (see for details) with the program MEGA6 (Tamura et al. Citation2013). The phylogenetic analysis supported the traditional taxonomy of the family Gramineae at the genus level. The species E. sibiricus Linn was found to be relatively closely related to Connorochloa tenuis compared with species of other genera in Gramineae (). We believe that it will provide valuable insight into conservation and evolutionary histories for this important species.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Dewey DR. 1984. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP (ed) Gene manipulation in plant improvement. New York: Plenum Press; pp.209–279.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lei Y, Zhao Y, Yu F, Li Y, Dou Q. 2014. Development and characterization of 53 polymorphic genomic-ssr markers in siberian wildrye (elymus sibiricus, l.). Conservation Genet Resour. 6:861–864.
- Löve A. 1982. Generic evolution of the wheatgrasses. Biologisches Zentralblatt. 101:199–212.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107:4623–4628.
- Motsny II, Simonenko VK. 1996. The influence of Elymus sibiricus L. genome on the diploidization system of wheat. Euphytica. 91:189–193.
- Nevski SA. 1934. Tribe Hordeae Benth. In: Komarov VL (ed) Flora of the USSR. Academy of Science Press USSR: Leningrad; p. 590–728.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang YJ, Ma PF, Li DZ. 2011. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae)). PLoS One. 6:e20596