Abstract
The Emei moustache toad, Leptobrachium boringii, is an IUCN red-listed vulnerable endemic precious species in China. In this study, we first report the nearly complete mitochondrial genome of L. boringii, which was 16,557 bp in length, containing 13 protein-coding genes, two rRNA genes, 22 transfer RNAs (tRNAs), and a putative control region. Phylogenetic analysis indicated that the L. boringii was clustered with L. leishanense. This work will provide a set of useful reference information on further molecular evolution studies of this endangered species.
As an endemic precious species of Megophryid toad, Leptobrachium boringii (Anura: Megophryidae) is mainly distributed in provinces of Sichuan, Hunan, Guizhou, Yunnan and Guangxi China (Wang and Zhao Citation1998, Liang and Guanfu Citation2004). L. boringii usually live in a favourable natural environment. However, for a long time, humans’ exploitation and demand is more than utilization and protection. Habitat environment have been dramatically damaged and now L. boringii has been classified as endangered by the IUCN Red List of Threatened Species (Liang and Guanfu Citation2004). Therefore, it is particularly important to take more reasonable and timely protection strategy. For better study and conservation of this species, herein, we first report the nearly complete mitochondrial genome of L. boringii to study its population and provide basic molecular data for protection of its wild resource.
The specimens of the tadpole of L. boringii were collected in Pengshui of Chongqing (107°58′E, 29°37′N) and preserved in 100% ethanol in the Museum of Chongqing Normal University until DNA extraction. The nearly complete genome sequence was 16,557 bp (GenBank: MH643882) and consisted of 13 protein-coding genes, two rRNA genes, 22 tRNA genes and one displacement loop (D-loop). Most genes were encoded on the H-strand, except the ND6 and seven tRNA genes. Over all base composition of the mtDNA was 27.7% A, 25.5% C, 15.2% G, 31.6% T. This mitogenome had two tRNAMet genes (tRNAMet1 and tRNAMet2 genes) derived from tandem duplication. The total length of the 13 protein-coding genes was 11,422 bp. Just like other megophrys, ten of them started with an ATG codon, while COX1, ATP8 and ND3 started with GTG. Stop codons were variable for all protein-coding genes. Four genes (ND2, COX1, ATP8 and ND4L) used complete stop codon TAA, and two genes (ND5 and ND6) ended with AGG, whereas other seven genes ended with incomplete stop codon (ND1, COX2, ATP6, COX3, ND3, ND4, CYTB ending with T) which may be presumably completed by post transcriptional polyadenylation with poly A tail (Ojala et al. Citation1981).
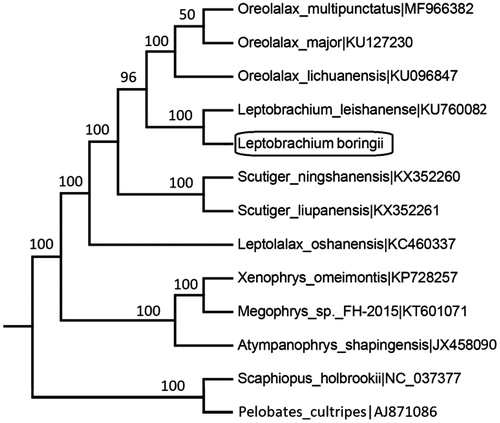
To ascertain the taxonomic position of L. boringii in family Megophryidae, a maximum likelihood (ML) phylogenetic analysis was constructed in PhyML 3.0 (Guindon and Gascuel Citation2003) based on the concatenated data set of 13 protein-coding genes (PCG) of other ten Megophryidae species, and two Pelobatidae species were used as outgroups. The result showed that the L. boringii was well grouped with L. leishanense (BP = 100) (), which the tree topology was congruent with traditional taxonomy.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Liang F, W. Guanfu (2004). Leptobrachium boringii. The IUCN Red List of Threatened Species 2004: e.T57625A11665713. http://dx.doi.org/10.2305/IUCN.UK.2004.RLTS.T57625A11665713.en.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Wang S, E. Zhao 1998. China Red Book of Endangered animals amphibia & reptilia. Beijing, Beijing Science Press.