Abstract
Abies kawakamii is endemic to the island of Taiwan and has been listed as a threatened species in the Red List. In present study, we reported the complete chloroplast genome of A. kawakamii. The chloroplast genome is 121,290 bp in size. It was composed of 114 genes and they were 68 peptide-encoding genes, 35 tRNA genes, four rRNA genes, six open reading frames and one pseudogene. Loss of ndh genes was identified in the genome of A. kawakamii. Inverted repeat sequences include trnS–psaM–ycf12–trnG and trnG–ycf12–psaM–trnS were recognized in 52-kb inversion points. The phylogenetic analysis confirms that the Abies species are strongly supported as monophyletic. The complete plastome of A. kawakamii will provide potential genetic resources for further conservation and management strategies.
Abies kawakamii (Hay.) Ito is endemic to the island of Taiwan and has been listed as a threatened species in the Red List (IUCN Citation2018). It is one of the southernmost firs (together with A. fansipanensis, native to Vietnam, and A. guatemalensis, from Mexico and Guatemala) (Liu Citation1971; Farjon Citation2001; Xiang et al. Citation2015). The Taiwan fir adapted to a cold, humid environment at high elevations and typically occurs in sheltered to windswept sites (Huang Citation2002). In the present study, we assembled and characterized the complete plastome of A. kawakamii. It will be fundamental to formulate conservation and management strategies for this threatened species.
We collected the plant material from the Hehuan Mountain of Taiwan. The voucher specimen (Chen C.-J., No. 9471) was deposited at the herbarium of Institute of Botany, CAS (PE). Complete chloroplast (cp) genome of Abies kawakamii was sequenced by HiSeq4000 of Illumina. Totally 10.3 million high-quality clean reads (150 bp PE read length) were obtained. In total, ca. 10.1 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and tRNAscan-SE v1.3.1 (Schattner et al. Citation2005) were used to align, assemble, and annotate the plastome.
The complete chloroplast genome consists of 121,290 bp for Abies kawakamii (GenBank: MH706726). The circle genome was comprised of a large single copy region (LSC with 65,646 bp), a small single copy region (SSC with 55,116 bp), and two inverted repeat regions (IR with 264 bp). The overall GC content of the A. kawakamii cp genome was 38.3%. It was composed of 114 genes and they were 68 peptide-encoding genes, 35 tRNA genes, four rRNA genes, six open reading frames and one pseudogene. All ndh genes have been lost in the genome of A. kawakamii. Short inverted repeat sequences were detected in 52-kb inversion points of the cp genome, which consist of trnS–psaM–ycf12–trnG and trnG–ycf12–psaM–trnS (1183 bp). Interestingly, such inverted repeats had been reported in several members of the genus Abies (A. koreana and A. ziyuanensis) (Yi et al. Citation2015; Shao et al. Citation2018).
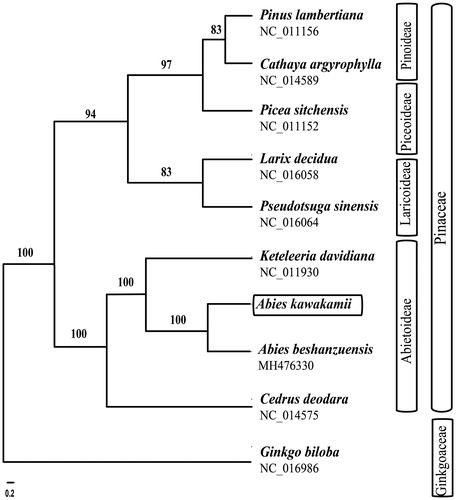
To infer the phylogenetic position of Abies kawakamii, nine chloroplast genomes were selected in Pinaceae with Ginkgo biloba (Ginkgoaceae) as the outgroup. These sequences were fully aligned with MAFFT v7.3 (Suita, Osaka, Japan) (Katoh and Standley Citation2013), and the maximum likelihood (ML) inference was performed using GTRþIþC model with RAxML v.8.2.1 (Karlsruhe, Germany) (Stamatakis Citation2014) on the CIPRES cluster service (Miller et al. Citation2010). The two Abies species (A. kawakamii and A. beshanenzuensis) were found to be a monophyletic group ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Farjon A. 2001. World checklist and bibliography of conifers, 2nd ed. London: The Royal Botanic Gardens, Kew.
- Huang KY. 2002. Evaluation of the topographic sheltering effects on the spatial pattern of Taiwan fir using aerial photography and GIS Int J Remote Sens. 23:2051–2069.
- IUCN. 2018. The IUCN red list of threatened species. Version. 2018-1; [accessed 2018 Aug 20]. www.iucnredlist.org.
- Liu TS. 1971. A monograph of the genus Abies. Taiwan: Department of Forestry, College of Agriculture, National Taiwan University.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE). New Orleans, LA: p. 1–8.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Shao YZ, Hu JT, Fan PZ, Liu YY, Wang YH. 2018. The complete chloroplast genome sequence of Abies beshanzuensis, a highly endangered fir species from south China. Mitochondrial DNA B. 3:923–924.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang XC. 2015. Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol Phylo and Evo. 82:1–14.
- Yi DK, Yang JC, So S, Joo MJ, Kim DK, Shin CH, Lee YM, Choi K. 2015. The complete plastid genome sequence of Abies koreana (Pinaceae: Abietoideae). Mitochondrial Dna. 27:1.