Abstract
An unknown species of marine sea lettuce was observed forming green tides consecutive years from 2014 to 2016 in Seaside, California. This Ulva sp. was similar in thallus size and shape to U. expansa. To confirm this identification, whole genome sequencing was performed on the bloom-forming species of Ulva and the holotype specimen of U. expansa. The complete green tide Ulva mitogenome is 64,143 bp in length, contains 65 genes, and displays high gene synteny with U. pertusa Kjellman. The mitogenome was incomplete for the holotype of U. expansa, but the analysis yielded the mitoexome, plastid, and nuclear genetic markers. These data verify that the native U. expansa is responsible for the blooms in central California.
Green tides are blooming events of green algae that result from eutrophication and warming seawater (Fletcher Citation1996, Shi & Wang Citation2009, Yoshida et al. Citation2015, Gao et al. Citation2017). They have been reported worldwide and their occurrences, particularly members of the genus Ulva, are on the rise (Smetacek & Zingone Citation2013). During the month of July in consecutive years 2014–2016, green tides of Ulva at Seaside, California were observed. To determine the identity of this Ulva species, genome sequencing was performed on a 2014 collection and on the holotype of U. expansa (Setchell) Setchell & N.L.Gardner (type locality: Monterey, California). Plastid markers were also sequenced from the 2015 and 2016 green tide collections to confirm their identities.
DNA was isolated from 5 × 5 mm2 snippets from the holotype specimen of U. expansa (specimen voucher UC 98481) and from a herbarium specimen of the bloom-forming Ulva from 2014 (UC 2050480) following the protocols of Lindstrom et al. (Citation2011) and Hughey and Gabrielson (Citation2012). The holotype was processed in 2016 using 76 bp paired-end Illumina library construction and sequencing, and the modern material in 2018 using 150 bp Illumina sequencing by myGenomics, LLC (Alpharetta, GA). The data were assembled using the default de novo settings in CLC Genomics Workbench 11 (®2018 CLC bio, a QIAGEN Company, Waltham, MA), and by mapping reads and contigs from the de novo analysis against the mitogenomes of U. linza, U. pertusa, and U. prolifera using default settings in Geneious R8 (Biomatters Limited, Auckland, New Zealand). The genes were annotated with Blastx and NCBI ORFfinder. The mitogenome data were aligned to other Ulva species with MAFFT (Katoh & Standley Citation2013). The RaxML analysis was executed using complete mitogenome sequences at Trex-online (Boc et al. Citation2012) with the GTR + gamma model and 1000 fast bootstraps, then visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
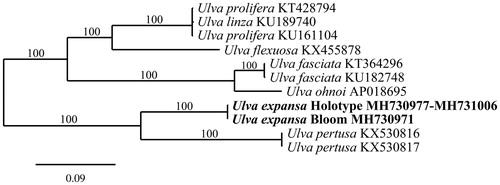
The complete mitogenome of the bloom-forming Ulva is 64,143 bp in length and contains 65 genes. The mitogenome of the holotype of U. expansa was incomplete; however, the genomic analysis yielded its mitoexome (GenBank numbers MH730977-MH731006). The two Ulva genomes contain cob, 2 rRNAs, 3 cox, 3 rpl, 5 ATP synthases, 7 orfs, 8 nad, 9 rps, and 27 tRNAs. Gene content, organization, and length of the blooming Ulva was similar to U. pertusa (Liu et al. Citation2017). Phylogenetic analysis of the two mitogenomes indicates a sister relationship to U. pertusa (). Analysis of rbcL sequences of the 2014–2016 bloom-forming Ulva from Seaside (GenBank numbers 2014 – MH730975, 2015 – MH730976, 2016 – MH746437) found three identical sequences, all differing from the holotype of U. expansa by only 2 bp. Comparison of plastid and nuclear markers of the holotype of U. expansa (GenBank numbers tufa – MH731007, UPA – MH731008, rbcL – MH731009; and SSU/ITS/LSU – MH730160) to the bloom-forming Ulva (GenBank numbers tufa – MH730973, UPA – MH730974, rbcL – MH730975; and SSU/ITS/LSU – MH730161) supports the conclusion that the bloom-forming Ulva from Seaside is the native central Californian species U. expansa.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Boc A, Diallo AB, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40:W573–W579.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Fletcher RL. 1996. The occurrence of “green tides” – a review. In: Schramm W, Nienhuis PH, editors. Marine benthic vegetation. Vol. 123, Ecological studies (analysis and synthesis). Berlin: Springer; p. 7–43.
- Gao G, Clare AS, Rose C, Caldwell GS. 2017. Eutrophication and warming-driven green tides (Ulva rigida) are predicted to increase under future climate change scenarios. Mar Pollut Bull. 114:439–447.
- Hughey JR, Gabrielson PW. 2012. Comment on “Acquiring DNA sequence data from dried archival red algae (Florideophyceae) for the purpose of applying available names to contemporary genetic species: a critical assessment.”. Botany. 90:1191–1194.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lindstrom SC, Hughey JR, Martone PT. 2011. New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific. Phycologia. 50:661–683.
- Liu F, Melton JT, Bi Y. 2017. Mitochondrial genomes of the green macroalga Ulva pertusa (Ulvophyceae, Chlorophyta): novel insights into the evolution of mitogenomes in the Ulvophyceae. J Phycol. 53:1010–1019.
- Shi W, Wang M. 2009. Green macroalgae blooms in the Yellow Sea during the spring and summer of 2008. J Geophys Res. 114:C12010.
- Smetacek V, Zingone A. 2013. Green and golden seaweed tides on the rise. Nature. 504:84–88.
- Yoshida G, Uchimura M, Hiraoka M. 2015. Persistent occurrence of floating Ulva green tide in Hiroshima Bay, Japan: seasonal succession and growth patterns of Ulva pertusa and Ulva spp. (Chlorophyta, Ulvales). Hydrobiologia. 758:223–233.