Abstract
The complete mitochondrial genome of Haemaphysalis japonica is reported for the first time in this study. Its entire mitogenome is 14,685 bp in length, containing 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and two non-coding regions. The phylogenetic analysis by Bayesian inference method show that H. japonica and other species of genus Haemaphysalis formed one clade, indicating that H. japonica belongs to the genus Haemaphysalis.
Ticks (Chelicerata: Anactinotrichida: Ixodida) are blood-feeding ectoparasites of terrestrial vertebrates (Chen et al. Citation2010; Burger et al. Citation2013). Haemaphysalis japonica (Ixodida: Ixodidae) is widely distributed in China, Russia, Germany, as well as temperate Eurasia, and Chinese-Russian border, respectively (Deng and Jiang Citation1991). Molecular characterizations of mitochondrial genomes have been made in some ticks (Guo et al. Citation2016; Chang et al. Citation2018), but H. japonica was not available for study.
The adult of H. japonica were collected by swiping flags on vegetations from a hill (39.782690 N, 115.956754 E), Fangshan district, Beijing, China, on 6 May 2017. The individual tick was stored in the Department of Parasitology, Heilongjiang Bayi Agricultural University (specimen no. BYNKPL-170506). Species identification was conducted by professor Sun Yi based on morphological features. Primers were designed for polymerase chain reaction (PCR) amplification and sequencing on the basis of the mitogenome sequence of Haemaphysalis concinna (GenBank accession no. NC_034785) (Chang et al. Citation2018).
The total length to the H. japonica mt genome was 14,685 bp (GenBank accession no. NC_037246), which contained 13 PCGs (cox1-3, nad1-6, nad4L, atp6, atp8, and cytb), two rRNA genes, 22 tRNA genes, and two NCR. The arrangement of the H. japonica was identical with that of hard ticks (Burger et al. Citation2013; Chang et al. Citation2018). The H. japonica mt genome encoded 3600 amino acids in total. The A + T content of PCGs ranged from 71.02% (cox1) to 84.57% (atp8). Codon usage analysis of H. japonica showed that ATG initiation codons were used in atp6, cox2, cox3, cytb, nad4, and nad4L, the others PCGs used ATT. Three kinds of termination codons (TAA, TAG, and T) were used. TAA was present in atp6, atp8, cox1, cox2, nad1, nad4L, nad5, and nad6, TAG was found in nad6 and cytb, and the others PCGs used the incomplete stop codon T. The incomplete stop codon was presumably completed as TAA by post transcriptional polyadenylation (Boore JL Citation1999). The size of the 22 tRNAs ranged from 54 to 67 bp. The rrnL (1189 bp) and rrnS (696 bp) were located between trnL2 and NCR1 and separated by trnV. The two NCR, long NCR (311 bp) and short NCR (308 bp), were located between rrnS-trnI and trnL1-trnC, which A + T contents were 68.79% and 67.74%, respectively.
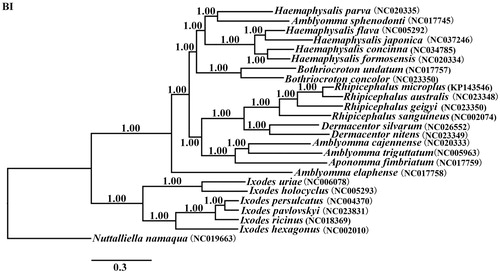
The concatenated amino acid sequences of 13 protein-coding genes were analyzed with Bayesian inference (BI), using Nuttalliellidae namaqua (NC_019663) as an outgroup. The result show that the tree divided into two large branches: Prostriata and Metastriata. Phylogenetic analysis revealed that the H. japonica and the others of genus Haemaphysalis are in the same clade with high statistical support, especially H. flava (). This result supports traditional taxonomic assignment of H. japonica to the genus Haemaphysalis. Amblyomma sphenodonti and Haemaphysalis parva are in the same branch, which is a controversial.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Burger TD, Shao R, Barker SC. 2013. Phylogenetic analysis of the mitochondrial genomes and nuclear rRNA genes of ticks reveals a deep phylogenetic structure within the genus Haemaphysalis and further elucidates the polyphyly of the genus Amblyomma with respect to Amblyomma sphenodonti and Amblyomma elaphense. Ticks Tick-Borne Dis. 4:265–274.
- Chang QC, Fu X, Song CL, Liu HB, Sun Y, Jia N, Jiang JF, Wang CR, Jiang BG. 2018. The complete mitochondrial genome of Haemaphysalis concinna (Ixodida: Ixodidae). Mitochondrial DNA Part B. 3:348–349.
- Chen Z, Yang XJ, Bu FJ, Yang XH, Yang XL, Liu JZ. 2010. Ticks (Acari: Ixodoidea: Argasidae, Ixodidae) of China. Exp Appl Acarol. 51:393–404.
- Deng GF, Jiang ZJ. 1991. Economic Insect Fauna of China. Fasc 39, Acari: Ixodidae. Beijing: Science Press, pp. 105–108.
- Guo DH, Zhang Y, Fu X, Gao Y, Liu YT, Qiu JH, Chang QC, Wang CR. 2016. Complete mitochondrial genomes of Dermacentor silvarum and comparative analyses with another hard tick Dermacentor nitens. Experimental Parasitology. 169:22–27.