Abstract
Abies ziyuanensis is critically endangered and endemic to Guangxi and Hunan provinces of China, with no more than 600 surviving individuals. In this study, we reported the complete chloroplast (cp) genome of A. ziyuanensis. The complete chloroplast genome is 121,274 bp in size. In total, 114 genes were identified, including 68 peptide-encoding genes, 35 tRNA genes, four rRNA genes, six open reading frames, and one pseudogene. Thirteen genes contain introns. In phylogenetic analysis, the tree supported the monophyly of the genus Abies. The complete plastome of A. ziyuanensis will provide potential genetic resources for further conservation and evolutionary studies of this highly endangered species.
Abies consists of about 50 species, which are widely distributed in the Northern Hemisphere with major centres of distribution in East Asia and America eastern Asia (Liu Citation1971; Farjon Citation2001). Abies are ecologically crucial for water and soil restoration in areas where they occur (Liu Citation1971). However, many firs have been listed as endangered species (Xiang et al. Citation2015). Abies ziyuanensis L.K. Fu & S.L. Mo is listed as highly endangered species in the Red List (IUCN Citation2018). Now Abies ziyuanensis is endemic to Guangxi and Hunan provinces of China with less than 600 surviving individuals. Here, we assembled and characterized the complete plastome of A. ziyuanensis. It will provide potential genetic resources for further conservation and evolutionary studies of A. ziyuanensis.
The plant material of A. ziyuanensis was collected from a single individual that lives in the natural forest habitat of Mt. Shunhuang (Hunan, China; 26.38°N, 111.01°E). Voucher specimen and DNA sample (Xiang Q.-P., No. ZY) were deposited in the herbarium of Institute of Botany, CAS (PE). In total, ca. 10.1 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and tRNAscan-SE version 1.3.1, (Santa Cruz, USA) (Schattner et al. Citation2005) were used to align, assemble, and annotate the plastome.
The full length of A. ziyuanensis chloroplast genome (GenBank Accession No. MH706705) was 121,274 bp and comprised of a large single copy region (LSC with 66,502 bp), a small single copy region (SSC with 54,244 bp), and two inverted repeat regions (IR with 264 bp). The overall GC content of the A. ziyuanensis cp genome was 38.30%. A total of 114 genes were contained in the cp genome (68 peptide-encoding genes, 35 tRNA genes, four rRNA genes, six open reading frames, and one pseudogene). Fifty-three protein coding, 16 tRNA genes, three open reading frames, and one pseudogene are located in the LSC region, while 15 protein-coding, 17 tRNA genes, 4 rRNA, and 3 open reading frames are located in the SSC region, respectively. Only one tRNA gene (trnI-CAU) is duplicated and located on the IR regions. All ndh genes have been lost in the genome of A. ziyuanensis like other cp genomes of family Pinaceae. Among the protein-coding genes, two genes (rps12 and ycf3) contained two introns, and other eleven genes (trnK-UUU, trnV-UAC, rpoC1, atpF, trnG-GCC, petB, petD, rpl16, rpl2, trnL-UAA, trnA-UGC) had one intron each. In previous studies, short inverted repeat sequences which consist of trnS-psaM-ycf12-trnG and trnG-ycf12-psaM-trnS (1183 bp) are located in 52-kb inversion points of the cp genome of A. ziyuanensis. Length and sequence of inverted repeats from A. ziyuanensis is identical with those of Abies koreana (Yi et al. Citation2015).
Twelve chloroplast genomes were selected to infer the phylogenetic relationships among the main representative species of Pinaceae with Ginkgo biloba (Ginkgoaceae) as the outgroup. These sequences were fully aligned with MAFFT version 7.3 (Suita, Osaka, Japan) (Katoh and Standley Citation2013), and the maximum likelihood (ML) inference was performed using GTRþIþC model with RAxML version 8.2.1 (Karlsruhe, Germany) (Stamatakis Citation2014) on the CIPRES cluster service (Miller et al. Citation2010). The result revealed that the four Abies species (A. ziyuanensis, A. koreana, Abiesneprolepis, and Abies beshanenzuensis) are strongly supported as monophyletic ().
Figure 1. The best maximum likelihood (ML) phylogram inferred from 12 chloroplast genomes in Pinaceae and Ginkgoaceae (bootstrap value are indicated on the branches).
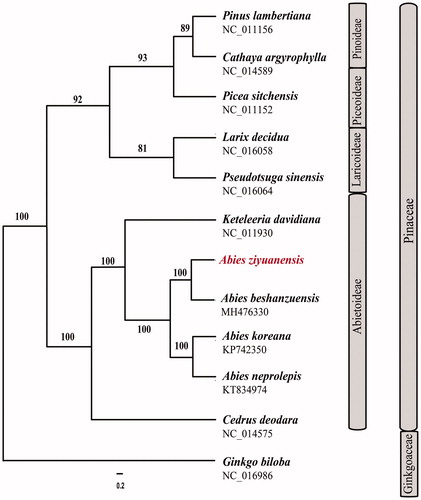
This study provides new insight into the cp genome evolution and phylogenetic relationships. Moreover, it would be fundamental to formulate potential conservation and management strategies for this endangered species in China.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Farjon A. 2001. World checklist and bibliography of conifers. 2nd ed. London: The Royal Botanic Gardens, Kew.
- IUCN. 2018. The IUCN red list of threatened species. Version 2018-1; [accessed 2018 Aug 20]. www.iucnredlist.org.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu TS. 1971. A monograph of the genus Abies. Taiwan, China: Department of Forestry, College of Agriculture, National Taiwan University.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the gateway computing environments workshop (GCE). Nov14–14; New Orleans (LA): IEEE. p. 1–8.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang XC. 2015. Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol Phylo Evo. 82:1–14.
- Yi DK, Yang JC, So S, Joo MJ, Kim DK, Shin CH, Lee YM, Choi K. 2015. The complete plastid genome sequence of Abies koreana (Pinaceae: Abietoideae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1.
