Abstract
The complete mitochondrial genome of Guillardia theta strain CCMP2712 was sequenced, assembled, and annotated in this study. The circular genome is 35,013 bp in size and it contains 36 protein-coding genes (PCGs), 28 transfer RNA genes (tRNA), and 2 ribosomal RNA genes (rRNA). The overall GC contents of the mitochondrial genome are 28.9%. The phylogenetic tree was constructed to validate the taxonomic relationship based on the complete mitogenomes of G. theta strain CCMP2712 through combining with seven Cryptophyta and four Heterokontophyta algae. The complete mitochondrial genome of the Guillardia theta strain CCMP2712 will provide more information for the evolution of secondary endosymbiotic species.
Guillardia theta, belonging to Cryptophyta, is considered as a transitional form in the secondary endosymbiotic, it is generally recognized to have photosynthesis ability through taking or engulfment photosynthetic eukaryotic algae (Curtis et al. Citation2012). Especially, there is a residual nuclear form (nucleomorph), which is considered to be a part of endosymbiont residual of the host nucleus in the chloroplasts outer membrane. Therefore, G. theta has four sets of genome, including nucleus, chloroplast, mitochondrial, and nucleomorph genomes, and at the same time, it can provide information for the persistence and evolution of eucaryotic organism and endosymbiont integration.
To reveal the fundamental issues of eukaryotic to eukaryotic symbiosis, the nucleomorph genomes, nuclear genomes (Curtis et al. 2012) and chloroplast genomes (Tang and Bi Citation2016) of Guillardia theta strain CCMP2712 have been sequenced, respectively. However, the sequence information of complete mitochondrial genome for G. theta has not been reported. There are still some limitations for the mitochondrial genome information of G. theta strain CCMP2712, although seven another Cryptophyta mitochondrial genomes have been published on the NCBI, including, Rhodomonas salina (GenBank accession number: AF288090) (Hauth et al. Citation2005), Hemiselmis andersenii CCMP644(NC010637) (Kim et al. Citation2008), Chroomonas placoidea strain CCAP978/8(MG680941), Cryptomonas curvata strain CNUKR(MG680942), Proteomonas sulcata strain CCMP705(MG680945), Storeatula sp. CCMP1868 (MG680943), and Teleaulax amphioxeia strain HACCP-CR01(MG680944) (Kim et al. Citation2018). In this study, the complete mitochondrial genome of the Guillardia theta strain CCMP2712 was recovered through Illumina sequencing data and then its genome profile and phyletic evolution were analyzed.
The sample of G. theta strain CCMP2712 was collected from Long Island Sound, North America (41.2264° N, 73.0639° W) and obtained from the Culture Collection of Algae at National Center for Marine Algae and Microbiota. The total genomic DNA of G. theta strain CCMP2712 was extracted by MiniBEST Plant Genomic DNA Extraction Kit (TaKaRa, Beijing, China) and deposited in our laboratory. Then, the purified DNA was built in a 270bp library and sequenced with Illumina HiSeq2500 platform by BGI Biotechnology Co. Ltd (Shenzhen, China). After the quality filtration of raw reads, the high-quality reads were assembled by SPAdes 3.9.0 (Bankevich et al. Citation2012). The one contig of the mitochondrial DNA was identified by sequence similarity to other Cryptophyta mitochondrial genomes. Finally, the mitochondrial genome annotation was performed by the MFannot tool and AEWEN Web server, and combined with manual correction.
This complete mitochondrial genome sequence together with gene annotations of G. theta CCMP2712 were submitted to GenBank database under the accession numbers of MH844545. The length of the mitochondrial genome of G. theta CCMP2712 is 35,013 bp with an intron sequence from 10630 to 10946, and it contains 36 protein-coding genes, 28 tRNA genes, and 2 rRNA genes. Meanwhile, 36 protein-coding genes include 5 atp genes, 3 cox genes, 9 nad genes, 15 ribosomal protein genes, cob and tatC genes. In the overall base composition of this genome, the percentage of GC is 28.9%. In addition, the circular mitochondrial genome map was drawn using Organellar Genome DRAW (OGDRAW) (Lohse et al. Citation2007).
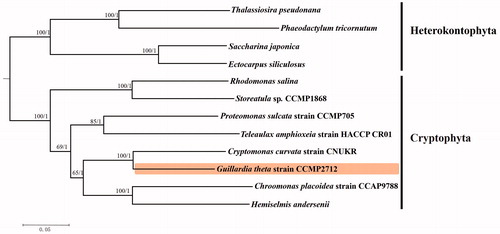
To validate the phylogenomic relationships, phylogenetic analysis was performed by using complete mitogenomes of seven Cryptophyta and four Heterokontophyta (Ectocarpus siliculosus [GenBank accession number: FP885846], Saccharina japonica [NC013476], Phaeodactylum tricornutum [NC016739], Thalassiosira pseudonana [NC007405]) algae species together with G. theta CCMP2712 mitochondrial genome sequence obtained in this study. Ultimately, 12 algae mitochondrial genomes were constructed by HomBlocks (Bi et al. Citation2017), resulting in alignments of each species with 5004 bp, including cob, cox1, nad5, and nad7 genes. For gene sequence analysis, the using Tbtools (Chen et al. Citation2018) were performed to gene sequence extraction and Local blast alignment. The phylogenetic tree was constructed based on Bayesian analysis (BI) and maximum likelihood (ML). At the same time, ML analysis was implemented in the soft RAxML version 7.0.3 (Stamatakis Citation2014), which relied on GTR + G+I model with 1000 bootstrap replicates. BI was performed by employing the soft MrBayes v.3.1.2 (Ronquist et al. Citation2012) for one million generations with the most appropriate model. The resulting consensus tree obtained from ML analysis was in accordance with those from BI. The phylogenetic tree showed that the G. theta strain CCMP2712 was clustered in the Cryptophyta clade and genetically close to Cryptomonas curvata strain CNUKR(MG680942) ().
Disclosure statement
The authors have declared that no competing interests exist. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SL, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2017. Homblocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Chen C, Xia R, Chen H, He Y. 2018. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv. 289660.
- Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, et al. 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 492:59
- Hauth AM, Maier UG, Lang BF, Gertraud B. 2005. The Rhodomonas salina mitochondrial genome: bacteria-like operons, compact gene arrangement and complex repeat region. Nucleic Acids Res. 33:4433–4442.
- Kim E, Lane CE, Curtis BA, Kozera C, Bowman S, Archibald JM. 2008. Complete sequence and analysis of the mitochondrial genome of Hemiselmis andersenii CCMP644 (Cryptophyceae). BMC Genomics. 9:215.
- Kim JI, Yoon HS, YiG SW, Archibald JM. 2018. Comparative mitochondrial genomics of cryptophyte algae: gene shuffling and dynamic mobile genetic elements. Bmc Genomics. 19:275.
- Lohse M, Drechsel O, Bock R. 2007. Organellargenomedraw (ogdraw): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 52:267–274.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tang X, Bi G. 2016. The complete chloroplast genome of Guillardia theta strain CCMP2712. Mitochondrial DNA A DNA Mapp Seq Anal. 27:4423–4424.