Abstract
Whole mitochondrial DNA (mtDNA) for Schizothorax esocinus sequence was found to be 16,591 bp with 13 protein-coding genes, 2 rRNAs genes, 22 tRNA genes, and 2 non-coding region. For estimating phylogenetic association of a species, protein coding genes of mitochondrial DNA are considered to be a great value. In this study, we sequenced protein-coding genes of Schizothorax esocinus, based upon which we compared the 23 Schizothoracinae fishes with S esocinus by phylogenetic association. We analyzed that S. esocinus to be closely associated with Schizothorax labiatus, Schizopyge niger, Schizothorax nepalensis with high-bootstrap values. Such empirical data would be important for understanding the phylogenetic relationship of Schizothorax species.
Introduction
Mitochondrial genome, because of its simple structure, abundant distribution, maternal inheritance, and its high mutation rate is thought to be the ideal marker for the genetic diversity in population studies, species identification and molecular phylogeny (Bucklin et al. Citation2011; Ortega-Ortiz et al. Citation2014). Genetic information in the form of mitochondrial DNA offers a base to manage and protect the diversity of biology and for the interpretation of evolutionary accounts of varied biological species it allows the researchers (Bernt et al. Citation2013). Self-replicating mitochondrial DNA of most of the animals is about 16-kb long; circular DNA molecule encodes for 13 protein-coding genes, (COI–III, Cytb b, ND1–6 and 4L, ATPase 6 and ATPase 8), 22 tRNAs, 2 rRNAs (12S, 16S), also having a control region that controls its replication and transcription, the gene content and organization of mitochondrial DNA is quite conserved, this conserved characteristics facilitates their identification and placement (Inoue et al. Citation2003; Peng et al. Citation2006). In order to infer the evolutionary relationship the fragments of DNA of 300–600 base pairs as of a single gene or a minor number of genes are regularly amplified and sequenced (Irwin et al. Citation1991). In inferences of phylogenetic and evolutionary relationships between diverse fish fauna, genome or gene based approaches have played a significant role (Karaiskou et al. Citation2003; Domingues et al. Citation2007). Next generation sequencing technologies in recent years have been used to achieve high coverage of large genomes in a cost-effective way (Schatz et al. Citation2010). For resolving phylogenetic trees branches, it is extensively known that information attained from a single gene is often inadequate (Sinclair et al. Citation2002; Afonso et al. Citation2011). So analysis of phylogenetics of Cyprinid taxa based on the functionally vital genes can help to understand the functional divergence and speciation (Kong et al. Citation2007). Majority of the whole mitochondrial genomic sequence occupied by 13 protein-coding genes. In the present study, we find out the phylogenetic relationship of Schizothorax esocinus with that of the other Schizothorax fishes on the basis of mitochondrial protein coding genes.
Materials and methods
Present study was conducted in Panjkora River, reflected as the chief life line of lower Dir, and part of Malakand division, situated in the province of Khyber Pakhtunkhwa, Pakistan. It lies in the range of Hindu Kush region between Latitude: 34° 39' 59.99" N, Longitude: 71° 45' 59.99" E The total area of Lower Dir region is 1,435,917 km2 having total population of 717,649 as per 2017 Census report. In the West of Swat it is surrounded by Afghanistan, from Malakand Agency on the South and Dir Upper on the North (Ahmad et al. Citation2014).
Fish sample collection
Fish samples of S. esocinus were collected from River Panjkora using different types of nets namely hand nets, cast nets and hooks. Transferred the specimen to the lab of Department of Zoology where the samples were stored. We preserved the tissue from muscles in 95% ethanol solution. Preserved samples then were moved to the lab for the DNA extraction.
Genomic DNA extraction, PCR and sequencing
For the amplification of the mtDNA of S. esocinus, we used standard high salt extraction method and extracted the mtDNA from the muscle tissues preserved in 95% ethanol (Miller et al. Citation1988). For polymerase chain reaction (PCR) amplification sixteen sets of primers were designed which were based on original mitochondrial genome DNA sequences of Cyprinid fish. Cocktail reaction included 25 µL to 6 µL of 10x buffer, 1.5 µL of every nucleotide (dNTP), 1 µL of every primer, about 1.5 µL of Taq DNA polymerase, 1–2 µL of template DNA. The process of thermocycling was started for 5 min at 94 °C, followed by 20 cycles at 94 °C for 30 s, 56 °C for 50 s, and 72 °C for 1 min 30 s, with the reduction of 0.1 °C for every cycle of annealing, with the annealing temperature at 54 °C. Then, followed by 12 more cycles, a final cycle of 8 min for extension was operated. On 1.2% of Agarose gel prepared in 1x Tris acetate-EDTA buffer, about 1 µl PCR product of all samples was electrophoresed at 80 V for 30 min, which was then stained with Ethidium bromide for visualization under ultra violet illumination in the Gel-Doc system. The PCR products purified through standard protocols were then sent for sequencing to the Sangon Biotech Company (Sangon Biotech Company Shanghai).
Analysis of Sequence
Using the program ClustalW and BioEdit (Hall Citation1999), the DNA sequences were aligned (Takamatsu et al. Citation1998). The DNA sequences was edited and analysed with Auto Assembler (Applied Bio systems) and DNASIS (Hitachi Software Engineering Co. Ltd). The locations of the 13 protein-codding genes were determined by comparisons of the amino acid sequences. The phylogenetic association was inferred by MEGA 6.0 (Tamura et al., Citation2013). For the phylogenetic association, the mtDNA, 13 sequenced protein coding genes of S. esocinus were used, and the sequences for infringing the phylogenetic relationships of S. esocinus with other 23 Schizothorax species, their sequences were retrieved from NCBI.
Results
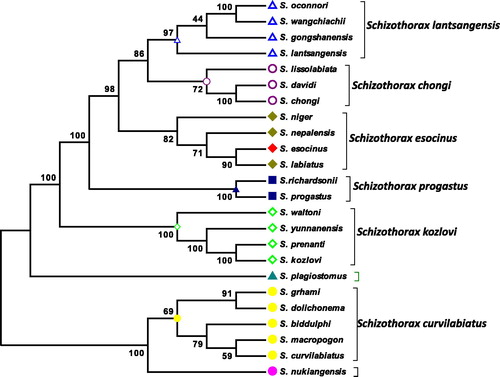
Combined data set of all the 13 protein-coding genes of 24 species of Schizothorax yielded that S. esocinus showed the closest relationship with the S. labiatus, Schizopyge niger, S. nepalensis, S. esocinus group appeared as the sister group with 100% posterior nodal probability with the S. progastus group and also showed the maximum probability with S. chongi group and S. lantsangensis group. Maximum bootstrap values support our results.
Discussion
Current study was conducted to find out the phylogenetic relationship of S. esocinus from Panjkora River, Khyber Pakhtunkhwa, Pakistan. The S. esocinus mitochondrial genome sequence was found to be 16,591 bp length. Like other Cyprinid fishes, genome of S. esocinus consisted of 13 protein coding gene (PCGs), 22 TRNAs gene, two ribosomal RNA (12S rRNAs and 16S rRNAs) genes, one control region, and light strand replication origin (OL) consisted of 33 bp. The neighbour joining analysis was performed in MEGA6 with 1000 bootstrap replicates (Yang Citation2007; Khan et al. Citation2016). Consequently, in inter-and intra-specific phylogeny in an animal’s mitochondrial genome the genetic information offered, in the studies consiquently (Qiu et al., Citation2011). Phylogenetic relationships among genera and species under Schizothoracinae have been investigated based on morphological characters, RAPD analysis. Cyprinid fishes phylogenetic analysis based on the functionally vital genes can support to know the functional divergence and speciation of these fishes. Consequently, it is the set of proteins which have the greatest marked variances between species observed, monophyly is intensely supported for the of Schizothorax fishes in the set of protein (Ficth and Ye Citation1991), and (Kong et al. Citation2007) Schizothorax esocinus showed the closest relationship with the S. labiatus, Schizopyge niger, S. nepalensis. Schizothorax esocinus group appeared as the sister group, with 100% posterior nodal probability, with the S. progastus group and also showed the maximum probability with S. chongi group and S. lantsangensis group. S. plagiostomus formed a different cluster. It was found that the species belonging to the northern Himalayas grouped together while species from north-eastern Himalayas remained separate (Barat et al. Citation2012). Maximum bootstrap values supports our results, these results are somewhat similar with the finding of Khan et al. (Citation2016). The phylogenetic relationship was inferred on the basis of whole mitochondrial data set.
Conclusion
The recent developments in molecular techniques based on genes are very much useful for establishing taxonomical and phylogenetic relationships among different species. The present study resolved the phylogenetic relationships of S. esocinus with the 24 species of subfamily Schizothoracinae. The study of phylogenetic relationship on the basis of sequences of protein-coding gene in these species offers useful visions to the phylogenetic status of Cyprinid fishes and provide the step for further investigations of issues with taxonomic and conservation and phylogenetic in this vital group of fishes.
Disclosure statement
The authors alone are responsible for the content and writing of the paper. The authors report no conflicts of interest.
References
- Afonso AS, Hazin FH, Carvalho F, Pacheco JC, Hazin H, Kerstetter DW, Murie D, Burgess GH. 2011. Fishing gear modifications to reduce elasmobranch mortality in pelagic and bottom longline fisheries off Northeast Brazil. Fish Res. 108(2): 336–343.
- Ahmad M, Rajapaksha AU, Lim JE, Zhang M, Bolan N, Mohan D, Vithanage M, Lee SS, Ok YS. 2014. Biochar as a sorbent for contaminant management in soil and water: a review. Chemosphere.99: 19–33.
- Barat A, Ali S, Sati J, Sivaraman G. 2012. Phylogenetic analysis of fishes of the subfamily Schizothoracinae (Teleostei: Cyprinidae) from Indian Himalayas using Cytochrome b gene. Indian J Fish. 59(1): 43–47.
- Bernt M, Braband A, Schierwater B, Stadler PF. 2013. Genetic aspects of mitochondrial genome evolution. Mol Phylogenet Evol. 69(2): 328–338.
- Bucklin A, Steinke D, Blanco-Bercial L. 2011. DNA barcoding of marine metazoa. Annu Rev Marine Sci. 3: 471–508.
- Domingues VS, Santos RS, Brito A, Alexandrou M, Almada VC. 2007. Mitochondrial and nuclear markers reveal isolation by distance and effects of Pleistocene glaciations in the northeastern Atlantic and Mediterranean populations of the white seabream (Diplodus sargus, L.). J Exp Marine Biol Ecol. 346(1):102–113.
- Ficth WM, Ye, J. 1991. Weighted parsimony: does it work. Phylogenetic analysis of DNA sequences (MM Miyamoto and J Cracraft, eds.). New York: Oxford Univ. Press, 147–154.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. London: Information Retrieval Ltd., c1979–c2000.
- Inoue JG, Miya M, Tsukamoto K, Nishida, M. 2003. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the “ancient fish”. Mol Phylogenet Evol. 26(1): 110–120.
- Irwin DM, Kocher TD, Wilson AC. 1991. Evolution of the cytochrome b gene of mammals. J Mol Evol. 32(2): 128–144.
- Karaiskou N, Apostolidis AP, Triantafyllidis A, Kouvatsi A, Triantaphyllidis C. 2003. Genetic identification and phylogeny of three species of the genus Trachurus based on mitochondrial DNA analysis. Marine Biotechnol. 5(5): 493–504.
- Khan M, Nasir Khan Khattak M, He D, Liang Y, Li C, Ullah Dawar F, Chen, Y. 2016. The mitochondrial genome of Schizothorax esocinus (Cypriniformes: Cyprinidae) from Northern Pakistan. Mit DNA Part A. 27(5): 3772–3773.
- Kong X, Wang X, Gan X, Li J, He, S. 2007. Phylogenetic relationships of Cyprinidae (Teleostei: Cypriniformes) inferred from the partial S6K1 gene sequences and implication of indel sites in intron 1. Sci China Life Sci. 50(6): 780–788.
- Miller S, Dykes D, Polesky H. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 16(3): 1215.
- Ortega-Ortiz CD, Elorriaga-Verplancken FR, Olivos-Ortiz A, Liñán-Cabello MA, Vargas-Bravo MH. 2014. Insights into the feeding habits of false killer whales (Pseudorca crassidens) in the Mexican Central Pacific. Aquat Mamm. 40(4): 386.
- Peng Z, Wang J, He S. 2006. The complete mitochondrial genome of the helmet catfish Cranoglanis bouderius (Siluriformes: Cranoglanididae) and the phylogeny of otophysan fishes. Gene. 376(2): 290–297.
- Qiu Y-W, Lin D, Liu J-Q, Zeng, EY. 2011. Bioaccumulation of trace metals in farmed fish from South China and potential risk assessment. Ecotoxicol Environ Safety. 74(3): 284–293.
- Schatz MC, Delcher AL, Salzberg SL. 2010. Assembly of large genomes using second-generation sequencing. Genome Res. 20(9): 1165–1173.
- Sinclair M, Arnason R, Csirke J, Karnicki Z, Sigurjonsson J, Skjoldal HR, Valdimarsson G. 2002. Responsible fisheries in the marine ecosystem. Fish Res. 58(3): 255–265.
- Takamatsu S, Hirata T, Sato Y. 1998. Phylogenetic analysis and predicted secondary structures of the rDNA internal transcribed spacers of the powdery mildew fungi (Erysiphaceae). Mycoscience. 39(4): 441–453.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol: mst197.
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8): 1586–1591.