Abstract
The complete chloroplast genome sequence of Cenchrus Purpureus, important silage in China, is presented in this article. The total genome size is 138,199 bp, containing a large single copy (LSC) region (81,161 bp) and a small single copy region (12,386 bp) which were separated by two inverted repeat (IRs) regions (22,326 bp). The overall GC contents of the plastid genome were 38.6%. In total, 136 unique genes were annotated and they were consisted of 87 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. Twenty-four genes duplicated in the LSC and IR regions. Eighteen genes contained one or two introns.
Cenchrus purpureus Schum., formerly Pennisetum purpureum (Chemisquy et al. Citation2010), originated in the subtropical Africa, is widely known as Elephant Grass, Napier Grass, or King Grass (Rueda et al. 2016). This giant grass has high biomass yield, and aroused the interest of many researchers for serving as a fodder for animal feeding (Valle et al. 2009; Chen et al. Citation2010), a wood substitute for the paper-mill industry (Madakadze et al. 2010), and a charcoal substitute for bioenergy production by direct biomass combustion (Samson et al. 2005; Morais et al. 2009).
The complete chloroplast genome of C. Purpureus was sequenced by MiSeq desktop sequencer of Illumina (San Diego, CA), assembled into the complete chloroplast genome by MITObim v1.8 (Hahn et al. Citation2013) with the reference chloroplast genome of C. americanus, annotated by Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004), and submitted to GenBank with the accession number of MF594682. The chloroplast DNA was extracted from a single individual C. Purpureus growing in germplasm garden of Sugarcane Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, China (22°51′27″N 108°15′25″E). DNA sample of C. Purpureus were stored in the laboratory of Sugarcane Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, China.
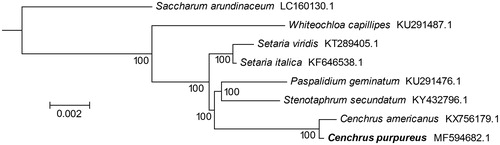
The total chloroplast genome size of C. Purpureus is 138,199 bp, containing a large single copy (LSC) region (81,161 bp) and a small single copy region (12,386 bp), which were separated by two inverted repeat (IRs) regions (22,326 bp). The overall GC contents of the plastid genome were 38.6%. In total, 136 unique genes were annotated, including 87 protein-coding genes, 8 rRNA genes, and 41 tRNA genes. There are 24 genes duplicated in the LSC and IR regions, including 9 protein-coding genes, 4 rRNA genes, and 11 tRNA genes. The protein-coding genes, rRNA genes, and tRNA genes account for 64.0%, 30.1%, and 5.9% of all annotated genes, respectively. Eighteen genes contained one or two introns, including the protein-coding genes, atpF, ndhA, ndhB, petB, rpl2, rps12, rps16, and ycf3. The Maximum Likelihood phylogenetic tree was generated using RAxML (Stamatakis Citation2014) based on the complete chloroplast genome of C. Purpureus and seven other species from the family Poaceae. The phylogenetic tree showed that C. Purpureus was closely related to C. americanus (). This published C. Purpureus chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Cenchrus and Poaceae.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Chemisquy MA, Giussani LM, Scataglini MA, Kellogg EA, Morrone O. 2010. Phylogenetic studies favour the unification of Pennisetum, Cenchrus and Odontelytrum (Poaceae): a combined nuclear, plastid and morphological analysis, and nomenclatural combinations in Cenchrus. Annals of Botany. 106:107–130.
- Chen ZT, He SL, Huang YB. 2010. Research progress of Pennisetum Rich. Acta Agrestia Sinica. 18:740–748.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Madakadze IC, Masamvu TM, Radiotis T, Li J, Smith DL. 2010. Evaluation of pulp and paper making characteristics of elephant grass (pennisetumpurpureumschum) and switchgrass (panicumvirgatumL.). Afr J Environ Sci Technol. 4:465–470.
- Morais R. F d, Souza B. J d, Leite JM, Soares L. H d B, Alves BJR, Boddey RM, Urquiaga S. 2009. Elephant grass genotypes for bioenergy production by direct biomass combustion. Pesq Agropec Bras. 44:133–140.
- Rueda JA, Ortega-Jiménez E, Hernández-Garay A, Enríquez-Quiroz JF, Guerrero-Rodríguez JD, Quero-Carrillo AR. 2016. Growth, yield, fiber content and lodging resistance in eight varieties of cenchruspurpureus, (schumach.) morrone intended as energy crop. Biomass Bioenergy. 88:59–65.
- Samson R, Mani S, Boddey R, Sokhansanj S, Quesada D, Urquiaga S, Reis V, Ho Lem C. 2005. The potential of C4 perennial grasses for developing a global BIOHEAT industry. Crit Rev Plant Sci. 24:461–495.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies . Bioinformatics. 30:1312–1313.
- Valle CBD, Jank L, Resende RMS. 2009. Tropical forage breeding in Brazil. Revista Ceres. 56:460–472.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.