Abstract
Dillenia indica Linn. is an endangered plant species occurring in southern Guangxi province and Yunnan province of China, which has been grown in gardens as an ornamental plant and is widely used in the medical field. Here, we report and characterize the complete plastid genome sequence of D. indica to provide genomic resources useful for promoting its conservation. The complete plastome is 159,266 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat (IR) regions of 26,457 bp, a large single-copy (LSC) region of 88,305 bp, and a small single-copy (SSC) region of 18,047 bp. The plastome contains 115 genes, consisting of 81 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA, and 16S rRNA). The overall A/T content in the plastome of D. indica is 63.40%. The complete plastome sequence of D. indica will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Dilleniaceae.
Dillenia indica Linn. (Dilleniaceae) is an evergreen tree grown in gardens as an ornamental plant (Kumar et al. Citation2011). It is a rare and Endangered plant of the family Dilleniaceae (Qin et al. Citation2017) and widely distributed in valleys and streamsides in southern Guangxi province and Yunnan province of China (Zhang and Klaus Citation2007). It is used for relieving abdominal pain and its fruit shows laxative properties (Kirtikar and Basu Citation2001). Dillenia indica was sampled from Baoting country in Hainan province of China (109.701°E, 18.642°N). A voucher specimen (Wang et al., B117) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Ampelopsis glandulosa (KT831767.1) (Raman and Park Citation2016) using MITO bim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Vitis aestivalis (KT997470.1). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of D. indica is found to possess a total length 159,266 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 26,457 bp, a large single-copy (LSC) region of 88,305 bp, and a small single-copy (SSC) region of 18,047 bp. The plastome contains 115 genes, consisting of 81 unique protein-coding genes (seven of which are duplicated in the IR: rps12, rps7, ndhB, ycf15, ycf2, rpl23, and rpl2), 30 unique tRNA genes (seven of which are duplicated in the IR: trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA and trnI-CAU), and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA, and 16S rRNA). Among these genes, 4 pseudogenes (ndhK, accD, rps19, and ycf1), 15 genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC, rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA) possessed a single intron and three genes (ycf3, clpP, and rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content in the plastome of D. indica is 63.40%, for which the corresponding value of the LSC, SSC and IR region were 65.80%, 69.80% and 57.20%, respectively.
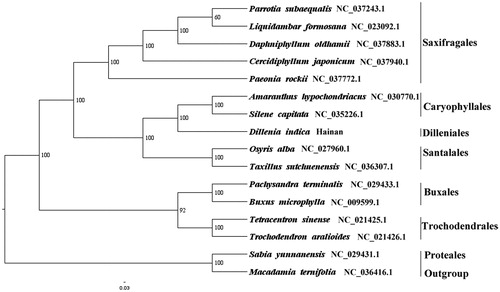
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 5 published complete plastomes of Saxifragales, 2 published complete plastomes of Caryophyllales, 2 published complete plastomes of Santalales, 2 published complete plastomes of Buxales, 2 published complete plastomes of Trochodendrales, using Sabia yunnanensis (Sabiaceae, Proteales) and Macadamia ternifolia (Proteaceae, Proteales) as outgroups. The phylogenetic analysis indicated that D. indica is closer to Caryophyllales than other orders in this study (). Most nodes in the plastome ML trees were strongly supported. The complete plastome sequence of D. indica will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Dilleniaceae.
Figure 1. The best ML phylogeny recovered from 16 complete plastome sequences by RAxML. Accession numbers: Dillenia indica Linn. (This study, GenBank Accession number: MH708162), Parrotia subaequalis NC_037243.1, Liquidambar formosana NC_023092.1, Daphniphyllum oldhamii NC_037883.1, Cercidiphyllum japonicum NC_037940.1, Paeonia rockii NC_037772.1, Amaranthus hypochondriacus NC_030770.1, Silene capitata NC_035226.1, Osyris alba NC_027960.1, Taxillus sutchuenensis NC_036307.1, Pachysandra terminalis NC_029433.1, Buxus microphylla NC_009599.1, Tetracentron sinense NC_021425.1, Trochodendron aralioides NC_021426.1; outgroups: Sabia yunnanensis NC_029431.1, Macadamia ternifolia NC_036416.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kirtikar KR, Basu BD. 2001. Indian medicinal plants. Dehradun: Oriental Enterprize.
- Kumar S, Kumar V, Prakash OM. 2011. Microscopic evaluation and physiochemical analysis of Dillenia indica leaf. Asian Pac J Trop Biomed. 1:337–340.
- Qin HN, Yang Y, Dong SY. 2017. A list of threatened species of higher plants in China. Biodiversity. 25: 697–744.
- Raman G, Park S. 2016. The complete chloroplast genome sequence of ampelopsis: gene organization, comparative analysis, and phylogenetic relationships to other angiosperms. Front Plant Sci. 7:341.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang ZX, Klaus K. 2007. Flora of China. Beijing: Science Press.
