Abstract
The mitochondrial genomes (mitogenomes) of ciliates are linear and relatively large (<80 kb). Until now, only 11 ciliate mitogenomes, either partial or complete, have been reported. The aim of the present study was to characterize the mitogenome of Pseudourostyla cristata (Ciliophora, Urostylida). The resulting mitogenome represents the first complete mitogenome for order Urostylida and is the largest sequenced ciliate mitogenome (∼76 kb), containing 31 protein-coding genes, 9 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and 21 unclassified open reading frames. Among the 11 ciliates whose mitogenomes have been identified, the mitogenome of P. cristata is most similar to that of Oxytricha trifallax, which is in the same subclass. However, the mitogenome of P. cristata is missing two genes (nad3 and nad6) and three split genes (nad4, nad5, and rpl6) that are found in the mitogenome of O. trifallax. Furthermore, the locations of the protein-coding and tRNA genes in P. cristata are different than those of the same genes in either Euplotes minuta or E. crassus, even though the species belong to the same class. This suggests that mitogenome structure is unlikely conserved in class level phylogenetic comparison.
Pseudourostyla cristata (Jerka-Dziadosz 1964) Borror, 1972 is a freshwater ciliate that was isolated from Mungyeong Saejae Provincial Park (36°46′45.77″N, 128°4′29.65″E), South Korea, in June 2014. Mitochondria were isolated from ∼200 live P. cristata specimens. Mitochondrial DNA (mtDNA) extraction, whole genome amplification, and mtDNA sequencing were performed according to Song et al. (Citation2016). DNA of specimen has been stored in the Korea Polar Research Institute (KOPRI), South Korea.
SOAP denovo, SSPACE 2.0 scaffolder, and Gap Closer tool were used (Boetzer and Pirovano Citation2014). The mitogenome was reconstructed using Geneious 6.1.3 (Biomatters, Ltd., Auckland, New Zealand) and the partial cytochrome b gene of P. cristata as an initial bait.
The complete mitogenome was annotated using MFANNOT (Beck and Lang Citation2010), and transfer RNA (tRNA) genes were identified using tRNAscan-SE1.2.1 (Lowe and Eddy Citation1997) with the default search mode. Gene locations were determined using the genoPlotR package in R (Guy et al. Citation2010).
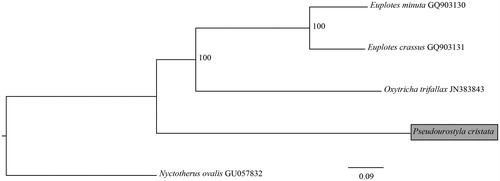
The phylogenetic analysis for mitogenome sequences was performed for the five ciliate species including our species (). Sequences were aligned using MAFFT (Katoh and Standley Citation2013). All gaps were removed using Gblocks 0.91b (Talavera and Castresana Citation2007). The phylogenetic relationships were reconstructed using the maximum likelihood method in the RAxML (Stamatakis Citation2006).
The resulting mitogenome of P. cristata represents the first complete mitogenome for order Urostylida and is the largest ciliate mitogenome (∼76 kb) sequenced to date (GenBank accession number: MH888186).
The mtDNA of P. cristata included 19 protein-coding genes with functions in the respiratory chain (complex I – nad1_a, 1_b, 2_a, 2_b, 4, 4L, 5, 7, 9, and 10; complex III – cob; complex IV – cox1, 2; complex V – atp9), small and large ribosomal subunit genes (rps2, 3_a, 3_b, 4, 7, 8, 10, 12, 13, 14, 19 and rpl2, 6, 14, and 16), 2 ribosomal RNA genes, 2 cytochrome c related genes, 9 tRNAs (trnE, trnF, trnH, trnK, trnL, trnM, trnQ, trnW, and trnY), and 21 unclassified open reading frames.
Mitogenomes have now been published for four spirotrich species, including P. cristata, O. trifallax, E. minuta, and E. crassus (Swart et al. Citation2011). The mitochondrial sequences of P. cristata and O. trifallax are longer and have relatively higher AT contents than those of E. minuta and E. crassus, as the genus Euplotes are phylogenetically close to our species (). Considering the identical genes between P. cristata and O. trifallax, the protein-coding gene composition and order of P. cristata mitogenome are similar to that of O. trifallax, except for the absence of nad3 and nad6 and no split genes of nad4, nad5, and rpl6.
Pseudourostyla cristata and O. trifallax, which belong to the same subclass, have relatively similar protein-coding gene sequences and locations, and E. minuta and E. crassus, which belong to the same genus, have identical protein-coding genes. Although there are not enough published ciliate mitogenomes to compare or analyze phylogenetically, more information of related species at the level of family as well as genus could be useful for more detailed study of mitogenome evolution and phylogenetic relationships in ciliates.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Beck N, Lang BF. 2010. MFannot, organelle genome annotation webserver. http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl.
- Boetzer M, Pirovano W. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinf. 15:211.
- Guy L, Kultima JR, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 26:2334–2335.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Song JH, Kim S, Shin S, Min GS. 2016. The complete mitochondrial genome of the mysid shrimp, Neomysis japonica (Crustacea, Malacostraca, Mysida). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2781–2782.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Swart EC, Nowacki M, Shum J, Stiles H, Higgins BP, Doak TG, Schotanus K, Magrini VJ, Minx P, Mardis ER, Landweber LF. 2011. The Oxytricha trifallax mitochondrial genome. Genome Biol Evol. 4:136–154.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577.