Abstract
Doubly uniparental inheritance (DUI) is a unique mode of mitochondrial (mt) inheritance found only in some bivalve molluscs. Under DUI, both maternal (i.e. female-transmitted or F-type) and paternal (i.e. male-transmitted or M-type) mitochondrial genomes are transmitted. In addition to typical mt genes, these F- and M-type genomes each contain a distinct novel open reading frame, termed the f- and m-orf, respectively. Herein, the M-type mitochondrial genomes of Lampsilis siliquoidea and Lampsilis powellii (two putative unionid species within Unionidae) are presented. Relatively few M-type mitochondrial genomes have been sequenced. Consequently, these results are valuable for the on-going study of patterns of molecular evolution of DUI genomes. Both a phylogenetic analysis and an in silico analysis of the novel ORFs were conducted. Consistent with previous studies of unionoid M-type genomes, our results demonstrate the presence of a putative transmembrane domain in these M-ORFs with properties similar to those of M-ORFs of closely related species.
Doubly uniparental inheritance (DUI) is a pattern of mitochondrial (mt) inheritance unique to Bivalvia (taxonomic distribution of DUI is reviewed in Gusman et al. Citation2016). Under DUI, both paternal and maternal mitochondria (and their corresponding male-transmitted or M-type and female-transmitted or F-type genomes) are transmitted within a species (reviewed in Breton et al. Citation2007; Passamonti and Ghiselli Citation2009; Zouros Citation2013). Unique open reading frames (ORFs) exist in each mitotype – termed f-orf and m-orf for the F- and M-type mtDNAs, respectively. These ORFs have been hypothesized to play a role in sex determination or sexual development in bivalves possesssing DUI (Breton et al. Citation2011). Previous in silico studies assessed these ORFs, and concluded that there are transmembrane domains (TMD) present within both the M- and F-ORF proteins, and that these TMDs may be variable in number and orientation (Mitchell et al. Citation2016; Guerra et al. Citation2017; Chase et al. Citation2018). Herein, whole M-type mitochondrial genome sequences are presented for two putative unionid species, i.e. the Fatmucket, Lampsilis siliquoidea, and the Arkansas Fatmucket, Lampsilis powellii. These data complement previous work on Lampsilis recently published by Robicheau et al. (Citation2018).
Lampsilis mussels were collected from Arkansas (Ouachita River, N34.56602, W094.02723; Poteau River, N34.89653, W94.39304), identified morphologically based on Harris et al. (Citation2004, Citation2010), and sexed by examining gonad tissue microscopically. DNA was extracted from male gonad using a QIAGEN DNeasy Blood and Tissue Kit. Complete mtDNAs were PCR amplified, sequenced, and assembled using the primers and methods of Breton et al. (Citation2011). Mitochondrial genomes were annotated using MITOS (Bernt et al. Citation2013). M-ORFs were identified using NCBI ORFfinder (National Centre for Biotechnology Information Citation2018). Both M-type mt genomes have been deposited in GenBank (accession codes MF326972 and MF326974).
Unionid M- and F-type mitochondrial genomes from GenBank were used to produce a phylogenetic tree with IQ-Tree (Nguyen et al. Citation2015). Protein-coding genes were extracted and aligned in Geneious v11.0.3 (Kearse et al. Citation2012) using ClustalW 2.1 (Thompson et al. Citation1994). A Maximum-likelihood tree of concatenated protein-coding genes was constructed with a Jukes–Cantor (JC) model. M-ORFs from Lampsilis M-type mitochondrial genomes were assessed in Geneious for transmembrane domains, and their orientation through the membrane was assessed using a hidden Markov model (Krogh et al. Citation2001).
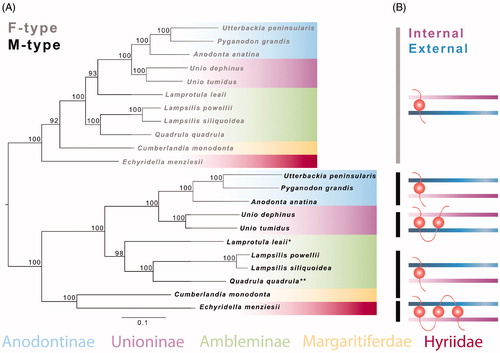
Our analysis produced a tree with separate F- and M-type lineages for gonochoristic members of the family Unionidae () as found in previous studies (Doucet-Beaupré et al. Citation2010; Gusman et al. Citation2016). Lampsilis powellii and L. siliquoidea M-ORFs putatively have one TMD with an external to internal orientation through the membrane (). Consequently, their number and orientation follow the same pattern as found in other unionids (Chase et al. Citation2018). More precisely, all unionoid F-ORFs have the same pattern (i.e. one TMD with an internal to external orientation), whereas M-ORFs present a variety of TMD characteristics and only closely related species (based on phylogeny presented in Lopes-Lima et al. Citation2017) share highly similar TMD patterns. Overall, these mitochondrial genomes will provide a valuable source of data for further studies of DUI, specifically relating to the under-represented M-types.
Figure 1. (A) Maximum-likelihood phylogeny using a Jukes-Cantor model and 1000 bootstrap replicates from 13 protein-coding mitochondrial genes from both F- and M-type mitochondrial DNA of unionid mussels. Bootstrap support values are reported at each node. (B) Orientation and number of transmembrane domains (TMD) for F-ORFs and M-ORFs of corresponding species in (A). Internal and external refer to orientation of the ORFs through the membrane, number of domains is represented by each circular structure. The mitotype, relative membrane location, and subfamily are denoted by color. *A defined M-ORF was not available. **Putative orientation could not be confidently determined. Phylogenetic analysis was conducted with IQ-Tree, and putative TMDs were assessed by Geneious using a hidden Markov model. GenBank accession numbers from top to bottom: HM856636, FJ809754, KF030964, KT326917, KY021078, JQ691662, MF326971, MF326973, FJ809750, KU873123, KU873121, HM856635, FJ809755, KF030963, KT326918, KY021073, KC847114, MF326972, MF326974, FJ809751, KU873124, and KU873122.
Acknowledgements
The following are thanked for providing specimens or assisting with collections: C.L. Davidson, D.M. Hayes, W.R. Posey II and J.H. Seagraves.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Breton S, Beaupré HD, Stewart DT, Hoeh WR, Blier PU. 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn’t one enough?. Trends Genet. 23:465–474.
- Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 28:1645–1659.
- Chase EE, Robicheau BM, Veinot S, Breton S, Stewart DT. 2018. The complete mitochondrial genome of the hermaphroditic freshwater mussel Anodonta cygnea (Bivalvia: Unionidae): in silico analyses of sex-specific ORFs across order Unionoida. BMC Genomics. 19:221.
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 10:50.
- Guerra D, Plazzi F, Stewart DT, Bogan AE, Hoeh WR, Breton S. 2017. Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida). Sci Rep. 7:1551.
- Gusman A, Lecomte S, Stewart DT, Passamonti M, Breton S. 2016. Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. PeerJ. 4:e2760.
- Harris JL, Hoeh WR, Christian AD, Walker J, Farris JL, Johnson RD, Gordon ME. 2004. Species limits and phylogeography of Lampsilinae (Bivalvia; Unionoida) in Arkansas with emphasis on species of Lampsilis. Final Report. Little Rock (AR): Arkansas Game and Fish Commission. p. 72.
- Harris JL, Posey WR, Davidson CL, Farris JL, Oetker SR, Stoeckel JN, Crump BG, Barnett MS, Martin HC, Matthews MW, et al. 2010. Unionoida (Mollusca: Margaritiferidae, Unionidae) in Arkansas, third status review. J Ark Acad Sci. 63:50–84.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 305:567–580.
- Lopes-Lima M, Froufe E, Do VT, Ghamizi M, Mock KE, Kebapçı Ü, Klishko O, Kovitvadhi S, Kovitvadhi U, Paulo OS, et al. 2017. Phylogeny of the most species-rich freshwater bivalve family (Bivalvia: Unionida: Unionidae): defining modern subfamilies and tribes. Mol Phylogenet Evol. 106:174–191.
- Mitchell A, Guerra D, Stewart D, Breton S. 2016. In silico analyses of mitochondrial ORFans in freshwater mussels (Bivalvia: Unionoida) provide a framework for future studies of their origin and function. BMC Genom. 17:597.
- National Centre for Biotechnology Information; ORFfinder. 2018. [accessed 2018 Jul 6]. https://www.ncbi.nlm.nih.gov/orffinder/.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Passamonti M, Ghiselli F. 2009. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 28:79–89.
- Robicheau BM, Chase EE, Hoeh WR, Harris JL, Stewart DT, Breton S. 2018. Evaluating the utility of the female-specific mitochondrial f-orf gene for population genetic, phylogeographic and systematic studies in freshwater mussels (Bivalvia: Unionida). PeerJ. 6:e5007.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680.
- Zouros E. 2013. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol Biol. 40:1–31.
