Abstract
The complete chloroplast genome sequence of Populus xiangchengensis C. Wang et Tung, an endemic species in southwest China, is presented in this study. The total genome size is 156,465 bp, with a typical quadripartite structure including a pair of inverted repeat (IRs, 27,570 bp) regions separated by a large single copy (LSC, 84,812 bp) region and a small single copy (SSC, 16,513 bp) region. The overall GC content of P. xiangchengensis plastid genome is 36.74%. The genome contains 130 genes, including 85 protein-coding genes, 37 transfer RNA genes, and eight ribosomal RNA genes. Among these genes, 12 genes have one intron and three genes contain two introns. The phylogenetic tree based on 26 complete plastomes of Salicaceae support close relationships among P. xiangchengensis, P. cathayana, and P. szechuanica.
The genus Populus in the family Salicaceae includes over 100 tree species distributed in the world, with 53 species in China. Populus species, collectively known as poplar, are known for their commercially and ecologically forest trees (Stettler et al. Citation1996; Hanzeh and Dayanandan Citation2004; Wan and Zhang Citation2013). Populus xiangchengesis, an endemic species, occurs only at high altitudes in Sichuan of SW China but is often confused with P. szechuanica, and the systematics of the species is unclear (Fang et al. Citation1999). For a better understanding of the relationships of P. xiangchengesis and P. szechuanica, it is necessary to reconstruct a phylogenetic tree of the Populus species based on high-throughput sequencing approaches.
The fresh leaves of P. xiangchengensis were sampled from Xiangcheng city (Sichuan, China; Coordinates: 99°40′33″E, 28°55′47″N; Altitude: 3530 m). Total genome DNA was extracted with the Ezup plant genomic DNA preps Kit (Sangon Biotech, Shanghai, China). The voucher specimens of P. xiangchengensis were deposited at the herbarium of Southwest Forestry University, and DNA samples were properly stored at Key Laboratory of State Forestry Administration on Biodiversity Conservation in Southwest China, Southwest Forestry University, Kunming, China. Total DNA was used to generate libraries with an average insert size of 400 bp and sequences using the Illumina Hiseq X platform. Approximately 15.0 GB of raw data were generated with 150 bp paired-end read lengths. Then the raw data was used to assemble the complete chloroplast genome using the software of GetOrganelle (Jin et al. Citation2018) with P. trichocarpa as the reference. Genome annotation was performed with the program Geneious R8 (Biomatters Ltd, Auckland, New Zealand), by comparing with the cp genome of P. trichocarpa. The cpDNA sequence with complete annation information was deposited at GenBank database under the accession number MH910611.
The plastome of P. xiangchengensis is a double-stranded circular DNA with the length of 156,465 bp, containing a large single copy (LSC) region of 84,812 bp, a small single copy (SSC) region of 16,513 bp, and a pair of inverted repeats of 27,570 bp. The GC content of the whole plastome is 36.74%, while the corresponding values of the LSC, SSC, and IR regions are 34.51%, 30.74%, and 41.97%, respectively. A total of 130 genes are annotated in the plastome, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among these genes, there are twelve genes (atpF, ndhA, ndhB, petB, rpoC1, rpl2, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) with one intron and three genes (ycf3, rps12, and clpP) with two introns. There are 18 genes duplicated in the IR regions, and 348 bp of 5′-rpl22 and 1689 bp of 3′-ycf1 are truncated at the boundaries of the IR regions.
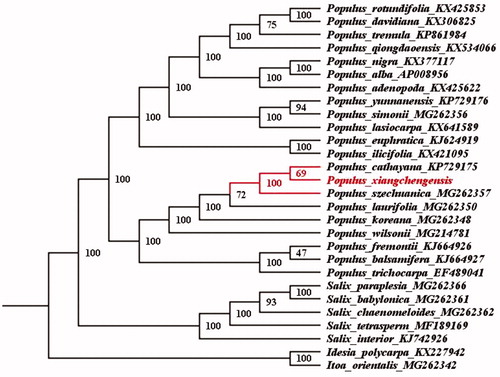
To determine the phylogenetic location of P. xiangchengensis with respect to the other Salicaceae with fully sequenced chloroplast genomes, the complete P. xiangchengensis plastome was used to reconstruct the phylogenetic relationships. With the plastomes of Idesia polycarpa and Itoa orientalis in the family of Flacourtiaceae as out-group (Yang et al. Citation2016), 26 chloroplast genome sequences of Salicaceae including P. adenopoda, P. alba, P. balsamifera, P. cathayana, P. davidiana, P. euphratica, P. fremontii, P. ilicifolia, P. koreana, P. lasiocarpa, P. laurifolia, P. nigra, P. qiongdaoensis, P. rotundifolia, P. simonii, P. szechuanica, P. tremula, P. trichocarpa, P. wilsonii, P. xiangchengensis, P. yunnanensis, Salix babylonica, S. chaenomeloides, S. interior, S. paraplesia, S. tetrasperm were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). A maximum likelihood method for phylogenetic analysis was performed base on GTR + I + G model in the RAxML version 8 program with 1000 bootstrap replicates (Darriba et al. Citation2012; Stamatakis Citation2014). Our chloroplast phylogenomic analysis confirms the monophyly of the Populus, in agreement with previously published phylogenetic studies (Hanzeh and Dayanandan Citation2004; Peng et al. Citation2005), and reveals that there are close relationships between P. xiangchengensis and P. cathayana rather than P. szechuanica ().
Data archiving statement
The plastome data of the P. xiangchengensis will be submitted to Genebank of NCBI through the revision process. The accession numbers from Genebank must be supplied before the final acceptance of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Fang ZF, Zhao SD, Skvortsov AK. 1999. Flora of China (English version). Beijing: Science Press.
- Hamzeh M, Dayanandan S. 2004. Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. Am J Bot. 91:1398–1408.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. BioRxiv. doi:10.1101/256479
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Peng YH, Lu ZX, Chen K, Luukkanen O, Korpelainen H, Li CY. 2005. Population genetic survey of Populus cathayana originating from southwestern Qinghai-Tibetan plateau of China based on SSR markers. Silvae Genetica. 54:116–122.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Stettler RF, Zsuffa L, Wu R. 1996. The role of hybridization in the genetic manipulation of Populus. In: Stettler RF, Bradshaw HD, Heilman PE, Hinckler TM, editors. Biology of Populus and its implications for management and conservation. Ottawa: Ganadian Government Publishing; 87–112.
- Wan XQ, Zhang F. 2013. An overview of Populus genetic resources in southwest China. Forestry Chronicle. 89:79–87.
- Yang WL, Wang WW, Zhang L, Chen ZY, Guo XY, Ma T. 2016. Characterization of the complete chloroplast genome of Idesia polycarpa. Conserv Genetics Res. 8:1–3.