Abstract
DNA barcode data of both endangered dolphin species, Platanista gangetica, and Orcaella brevirostris are lacking in the global database. The study generated partial mitochondrial cytochrome c oxidase subunit I (mtCOI) gene sequences of the two highly threatened dolphin species from India. The family Platanistidae with one species and family Delphinidae with 12 species were showed 1.2%–19.7% between species genetic divergence in the studied dataset. Both P. gangetica and O. brevirostris were revealed distinct clade in neighbor-joining (NJ) phylogeny. Further, D. delphis, S. coeruleoalba, and T. truncatus have shown more than one clade and comparatively high genetic divergence within the species suggested prominent genetic variation between different populations. The study recommends generating more DNA barcode data from diverse habitats of P. gangetica and O. brevirostris to evaluate the exact population structure and adopting better conservation strategies thereafter.
1. Introduction
Dolphins are one of the oldest widely distributed aquatic mammal and are classified under cetaceans (Arnason et al. Citation2004; Zhou et al. Citation2011). The majority of extant dolphin species are known from marine eco-system, while few species are distributed in freshwater riverine (Hamilton et al. Citation2001). The animal has a streamlined body with one pair of limbs that are modified into flippers, a tail fin, and spherical heads. Although the distribution patterns of dolphins are widespread, many of them prefer warmer waters of the tropical zones. Dolphins are frequently leaped above the water surface and their charismatic behavior occasionally acts as recreational purposes. The South Asian river dolphin, Platanista gangetica, locally known as Susu; is regarded as ‘National Aquatic Animal’ of India in 2009. The species is endemic to Bangladesh, India, Nepal, and Pakistan (Shostell and García Citation2010). The anecdotal reports often baffled the identity of two populations of P. gangetica from river Ganges and Indus (Sinha et al. Citation2010). Besides, the Irrawaddy dolphin, Orcaella brevirostris generally inhabits inshore coastal habitats and deltas of riverine systems in South and Southeast Asia (James et al. Citation1989). The occurrence of this species was known from Bangladesh, Brunei, Cambodia, India, Indonesia, Laos, Malaysia, Myanmar, Philippines, Singapore, Thailand, and Viet Nam. Both P. gangetica and O. brevirostris are playing an important role to balance the eco-system, fighting infectious diseases among fish populations and well-known as an indicator species in both marine and freshwater environments (Sinha and Sharma Citation2003).
However, the population of dolphin species is dramatically declining in recent past due to habitat loss (Paudel et al. Citation2015). Besides the environmental calamity, the water development projects, pollution, noise from vessel traffic, indocile tourism, poaching for bushmeat and traditional medicine, and unintentional killing through entanglement in fishing gear, the dolphin population are dropping down throughout the world including in India (Dhandapani Citation1997; Sahu et al. Citation1998). Both P. gangetica and O. brevirostris are categorized as ‘endangered’ in the IUCN red list of threatened species and ‘Schedule I’ in Indian Wildlife (Protection) Act, 1972 (IUCN Citation2018). Further, to mitigate the threats, several conservation agencies developed several plans of action and scientific research activities to fill the dearth in different eco-biological aspects and protect this species in their habitats. However, the contemporary DNA barcode-based scientific intervention is lacking for this species. In recent past DNA barcoding technique bloomed as a supplementary tool in taxonomic research. The efficacy of mitochondrial marker (COI) has been evidenced for species identification and systematics research (Tyagi et al. Citation2017). Therefore, the present study was aimed to generate DNA barcode data of two dolphin species from India. The study further tested the efficacy of the DNA barcoding region to differentiate the aimed taxa with other dolphin species.
2. Materials and methods
2.1. Collection of confiscated sample
The biological samples (raw tissue) were received at Centre for DNA Taxonomy (CDT) laboratory, Zoological Survey of India (ZSI), Kolkata from the two regional centres of ZSI. The tissue sample was collected from dead P. gangetica by Department of Environment and Forests, Government of Bihar from river Ganges (25.58 N 85.25 E) on 7 November 2017 and donated to the Museum, Gangetic Plains Regional Centre (GPRC), ZSI. The tissue sample of O. brevirostris was collected by the Marine Aquarium and Regional Centre (MARC) on 23 February 2018 from Digha, Gangadharpur, West Bengal (21.62 N 87.53 E). The subsamples were made from each of the samples for DNA analysis and immediately washing 3–4 times by 70% molecular grade ethanol and nuclease-free water. While subsampling, the surface layers of the tissue samples were avoided by using a sterile surgical blade in a separate room to nullify the contamination. After DNA isolation, the tissue samples were stored in –80 °C at CDT, ZSI, Kolkata.
2.2. DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted by using the QIAamp DNA Investigator Kit (QIAGEN Inc., Germantown, MD) as per the standard protocol. The genomic DNA was checked by 1% Agarose gel and quantification of the extracted DNA was determined by using the Nanodrop (Eppendorf). The published primer pairs were used to amplify the partial mtCOI segment of the mitochondrial gene in a Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA) (Ward et al. Citation2005). The 25µl PCR mixture contains 10 pmol of each primer, 20 ng of DNA template, 1X PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 0.25 U of Platinum Taq DNA Polymerase High fidelity (Invitrogen). The PCR products were checked in 1% agarose gel containing ethidium bromide (10 mg/ml). Further, the PCR products were purified using the QIAquickR Gel extraction kit (QIAGEN Inc., Germantown MD), and cycle sequencing products were cleaned by using standard BigDye X Terminator Purification Kit (Applied Biosystems, Foster City, CA). Sequencing was done bi-directionally in 48 capillary arrays 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) following Sanger sequencing methods in the in-house sequencing facilities in ZSI, Kolkata.
2.3. Sequence check and dataset preparation
The generated sequences were checked by Sequence Analysis software (ABI) and assured by the online nucleotide BLAST program and ORF finder to examine the complete alignment and start-stop codon (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Finally, the generated sequences were submitted in the GenBank database to acquire the specific accession number. The mtCOI sequences were identified in the online identification system, in GenBank with ‘highly similar sequences (Megablast)’. Further, 70 publicly available mtCOI sequences of same and related species were acquired from GenBank. Total 72 sequences were aligned using ClustalX software to form a combined dataset (Thompson et al. Citation1997). To infer the genetic distance and clustering pattern of the studied taxa, the dataset was analyzed through neighbor-joining (NJ) tree and Kimura 2 parameter (K2P) by using MEGA6 (Tamura et al. Citation2013).
3. Results and discussion
Since the period 2011–2020 has been declared the biodiversity decade, the Convention of Biodiversity (CBD) has laid major emphasis on the inventory and conservation of biodiversity. Thus, inventorying and monitoring of the biotic components are important for the conservation of the threatened species and management of their ecosystem. The traditional taxonomic practices, particularly in the cases of organisms having morphological plasticity sometime furnished wrong information leading to the disintegration of a single species into multiple species. Similarly, the organisms having static morphology have been ignored in taxonomic scrutiny; resultantly several genetically distant taxa are considered in one species or cryptic complex. Thus, improvement of identification systems in biodiversity research is the need of the hour.
Conservation of both Gangetic and Irrawaddy dolphins are no way behind any importance in Indian waters (Reeves et al. Citation2004). It is reported that within the long stretch of River Ganges and Brahmaputra, the population of P. gangetica is segregated into different subpopulations at different sporadic locations and the O. brevirostris population is restricted in a single largest estuarine habitat in Chilika Lake in eastern India (Smith et al. Citation2008). To develop the precise conservation management of both dolphins, the generation of molecular data is compulsory. As of now several molecular markers, cytochrome b gene, 12S, and 16S ribosomal RNA, as well as A + T rich control regions have been studied to corroborate the species identification (Hrbek et al. Citation2014), population estimation (Jayasankar et al. Citation2011; Bayas-Rea et al. Citation2018), understand the mechanisms leading to speciation in the context of gene flow (Amaral et al. Citation2014), and evolutionary pattern (Arnason and Gullberg Citation1996; Nikaido et al. Citation2001; Verma et al. Citation2004; McGowen Citation2011) and range distribution (Farias-Curtidor et al. Citation2017) of dolphins throughout the world.
Till date, DNA barcodes of P. gangetica and O. brevirostris are unknown to the world. To fulfill the gap in the global database, the study generated first mtCOI data of two dolphin species from India and submitted in GenBank. The O. brevirostris (ZSI_D1 MH844860) and P. gangetica (ZSI_D2 MH844861) shows 99% identity with their respective complete mitochondrial genome sequences by BLASTn sequence similarity search in the GenBank database. The family Platanistidae with one species and family Delphinidae with 12 species shows 6.9% overall mean genetic divergence in the studied dataset (). The highest within species genetic divergence (6.57%) resulted in D. delphis while the other two species S. coeruleoalba and T. truncatus shows 1.16% and 3.16% within species genetic divergence respectively (). The genetic divergence between the species ranged from 1.2% to 19.7% in the studied dataset. Further, the family Delphinidae shows 9.9% highest genetic divergence between the species. Most of the studied species were clustered separately in the NJ phylogeny (). The species with high genetic divergence, D. delphis, S. coeruleoalba, and T. truncates shows more than one clade in the NJ phylogeny, correlate to the different populations (). The studied DNA data would be helpful for species identification through similarity search in a global database where the morphological data are missing. Further, we recommend generation of more DNA barcode data from widespread locations of P. gangetica and O. brevirostris to evaluate the exact population structure for adopting better conservation strategies.
Figure 1. (A) Dead sample of Irrawaddy dolphin, Orcaella brevirostris in Digha sea beach, West Bengal. (B) Neighbour-joining (NJ) tree of the studied freshwater and marine dolphins with bootstrap support. Three database sequences of the whale species, Balaena mysticetus used as an out-group in the phylogeny. The species under family Delphinidae and Platanistidae are clustered together and represent by color bars. The ambiguous clades of the studied species revealed in NJ phylogeny marked by red, blue and brown colors boxes. The line drawings of the dolphin species were acquired from the free media repository of Wikimedia Commons (https://commons.wikimedia.org) and are superimposed in the phylogeny.
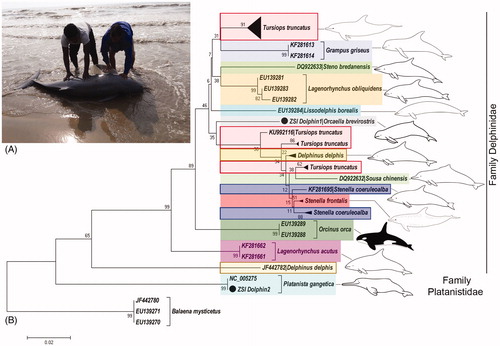
Table 1. The K2P genetic divergence of the studied freshwater and marine dolphins.
Acknowledgements
We thank the Director, Zoological Survey of India, Ministry of Environment, Forest and Climate Change (MoEF&CC) for providing necessary working facilities.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Amaral AR, Lovewell G, Coelho MM, Amato G, Rosenbaum HC. 2014. Hybrid speciation in a marine mammal: the Clymene Dolphin (Stenella clymene). PLoS One. 9:e83645.
- Arnason U, Gullberg A. 1996. Cytochrome b mucleotide sequences and the identification of five primary lineages of extant Cetaceans. Mol Biol Evol. 13:407–417.
- Arnason U, Gullberg A, Janke A. 2004. Mitogenomic analyses provide new insights into cetacean origin and evolution. Gene. 333:27–34.
- Bayas-Rea RA, Felix F, Montufar R. 2018. Genetic divergence and fine scale population structure of the common bottlenose dolphin (Tursiops truncatus, Montagu) found in the Gulf of Guayaquil, Ecuador. PeerJ. 6:e4589.
- Braulik GT, Barnett R, Odon V, Islas-Villanueva V, Hoelzel AR, Graves JA. 2014. One species or two? Vicariance, lineage divergence and low mtDNA diversity in geographically isolated populations of South Asian River Dolphin. J Mamm Evol. 22:111–120.
- Dhandapani P. 1997. The conservation of the potentially endangered Irrawaddy River dolphin Orcaella brevirostris in Chilka Lagoon, Orissa, India. J Bombay Nat Hist Soc. 94:536–539.
- Farías-Curtidor N, Barragán-Barrera DC, Chávez-Carreño PA, Jiménez-Pinedo C, Palacios DM, Caicedo D, Trujillo F, Caballero S. 2017. Range extension for the common dolphin (Delphinus sp.) to the Colombian Caribbean, with taxonomic implications from genetic barcoding and phylogenetic analyses. PLoS One. 12:e0171000.
- Hamilton H, Caballero S, Collins AG, Brownell JRL. 2001. Evolution of river dolphins. Proc Biol Sci. 268:549–556.
- Hrbek T, da Silva VMF, Dutra N, Gravena W, Martin AR, Farias IP. 2014. A new species of River Dolphin from Brazil or: how little do we know our biodiversity. PLoS One. 9:e83623.
- IUCN. 2018. The IUCN red list of threatened species, Version 2018.1. https://www.iucnredlist.org.
- James PSBR, Rajgopalan M, Dan SS, Fernando AB, Selvaraj V. 1989. On the mortality and stranding of marine mammals and turtles at Gahirmatha, Orissa from 1983 to 1987. J Mar Biol Ass India. 31:28–35.
- Jayasankar P, Patel A, Khan M, Das P, Panda S. 2011. Mitochondrial DNA diversity and PCR-based sex determination of Irrawaddy dolphin (Orcaella brevirostris) from Chilika Lagoon, India. Mol Biol Rep. 38:1661–1668.
- McGowen MR. 2011. Toward the resolution of an explosive radiation-a multilocus phylogeny of oceanic dolphins (Delphinidae)). Mol Phylogenet Evol. 60:345–357.
- Nikaido M, Matsuno F, Hamilton H, Brownell RL, Cao Y, Ding W, Zuoyan Z, Shedlock AM, Fordyce RE, Hasegawa M, et al. 2001. Retroposon analysis of major cetacean lineages: the monophyly of toothed whales and the paraphyly of river dolphins. Proc Natl Acad Sci USA. 98:7384–7389.
- Paudel S, Pal P, Cove MV, Jnawali SR, Abel G, Koprowski JL, Ranabhat R. 2015. The endangered Ganges River dolphin Platanista gangetica gangetica in Nepal: abundance, habitat and conservation threats. Endanger Species Res. 29:59–68.
- Reeves RR, Perrin WF, Taylor BL, Baker CS, Mesnick SL. 2004. Report of the workshop on the shortcomings of cetacean taxonomy in relation to the needs of conservation and management. La Jolla, California: NOAA.
- Sahu HK, Kar SK, Patnaik SK. 1998. Study on some aspects of Irrawaddy river dolphin Orcaella brevirostris gray in Chilika Lake, Orissa. Indian Forum. 24:803–809.
- Shostell JM, García MR. 2010. An introduction to river dolphin species. In: Ruiz-Garcia M, Shostell JM, editors. Biology, evolution and conservation of river dolphins within South America and Asia. New York (NY): NOVA Science Publishers; p.1–28.
- Sinha RK, Behera S, Choudhary BC. 2010. Conservation action plan for the Gangetic river dolphin 2012-2020. India: National Ganga River Basin Authority, Ministry of Environment and Forests.
- Sinha RK, Sharma G. 2003. Current status of the Ganges River dolphin, Platanista gangetica in the rivers Kosi and Son, Bihar, India. J Bombay Nat Hist Soc. 100:27–37.
- Smith BD, Ahmed B, Mowgli RM, Strindberg S. 2008. Species occurrence and distributional ecology of nearshore cetaceans in the Bay of Bengal, Bangladesh, with abundance estimates for Irrawaddy dolphins Orcaella brevisrostris and finless porpoise Neophocaena phocaenoides. J Cetacean Res Manag. 10:45–58.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Tyagi K, Kumar V, Singha D, Chandra K, Laskar BA, Kundu S, et al. 2017. DNA barcoding studies on Thrips in India: cryptic species, species complexes. Scientific Rep. 7:1–14.
- Verma S, Sinha R, Singh L. 2004. Phylogetic position of Platanista gangetica: insights from the mitochondrial cytochrome b and nuclear interphotoreceptor retinoid-binding protein gene sequences. Mol Phylogenet Evol. 33:280–288.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding of Australia’s fish species. Philos Trans Royal Soc B: Biol Sci. 360:1847–1857.
- Zhou X, Xu S, Yang Y, Zhou K, Yang G. 2011. Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol Phylogenet Evol. 61:255–264.
