Abstract
In this study, we first determined and described the complete mitochondrial genome of Styracaster yapensis, a member of the family Porcellanasteridae. The complete mitogenome of S. yapensis is 16360 bp in length, containing 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and one control region. Phylogenetic analysis indicated that S. yapensis was closely related to two other Paxillosida species.
The starfish species of the genus Styracaster (Paxillosida: Porcellanasteridae) are mud bottom dwellers, and living at abyssal and hadal depths in the world ocean (Madsen Citation1961; Mironov et al. Citation2016; Zhang et al. Citation2017). Styracaster yapensis was first reported and described by Zhang et al. (Citation2017), but there is little molecular information about S. yapensis available. Here we first sequenced and characterized the complete mitochondrial genome of S. yapensis, and examined the phylogenetic position of S. yapensis within Paxillosida.
The specimen of S. yapensis was collected at Mariana Trench, in July 2016 (10°53.3210′N, 142°13.6523′E, at 6300 m depth), and preserved in 95% ethanol. Total genomic DNA was extracted from muscle tissue and stored at –80 °C in Institute of Deep-sea Science and Engineering, CAS. The complete mitochondrial genome of S. yapensis was sequenced using the long-PCR technique. The annotation of mitochondrial protein-coding genes and ribosomal RNA genes were identified using DOGMA (Wyman et al. Citation2004). The online automated program MITOS webserver (http://mitos.bioinf.uni-leipzig.de/index.py) was used to predict the transfer RNA genes (Bernt et al. Citation2013). The S. yapensis mitogenome has been deposited in the GenBank database (accession number: MH648613).
The mitogenome sequence is 16360 bp in length, containing 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and one control region. The overall nucleotide composition is 34.36% A, 22.50% C, 11.55% G, 31.58% T, with AT content of 65.95%. Fifteen of the genes are encoded on the light strand (L-strand), while the other 22 on the heavy strand (H-strand).
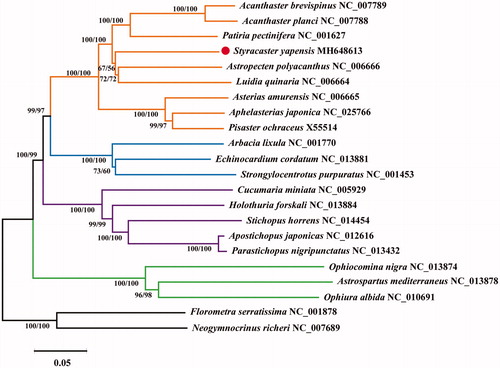
Twenty-two echinoderm mitogenomes including the one obtained in this study were used for phylogenetic analysis. Florometra serratissima and Neogymnocrinus richeri were used as an outgroup. Phylogenetic reconstruction was performed using neighbor-joining (NJ) and maximum likelihood (ML) with MEGA 5.0 (Tamura et al. Citation2011). The assessments of node reliability in the NJ and ML analyses were done by using 1,000 bootstrap replicates. Phylogenetic analyses showed that S. yapensis was closely related to Astropecten polyacanthus and Luidia quinaria. These three starfish formed the Paxillosida clade. Within superorder Valvatacea, S. yapensis (Paxillosida) was sister to species in the order Valvatida (). This study will provide valuable molecular data for further evolutionary analysis of this deep-sea species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Madsen FJ. 1961. The Porcellanasteridae. A monographic revision of an abyssal group of sea-stars. Galathea Rep. 4:33–174.
- Mironov AN, Dilman AB, Vladychenskaya IP, Petrov NB. 2016. Adaptive strategy of the porcellanasterid sea stars. Biol Bullet. 43:503–516.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang RY, Zhou YD, Lu B, Wang CS. 2017. A new species in the genus Styracaster (Echinodermata: Asteroidea: Porcellanasteridae) from hadal depth of the Yap Trench in the western Pacific. Zootaxa. 4338:153–162.