Abstract
The complete mitochondrial genome of this species was first determined in this study. We describe the complete mitogenome of Stolephorus insularis in this study. The mitogenome is 16711 nucleotides long and contains 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, and two main non-coding regions. The overall base composition includes A (28.0%), C (26.8%), T (27.8%), and G (17.4%). Furthermore, the 13 PCGs encode 3624 amino acids in total. All the PCGs use the initiation codon ATG except COI uses GTG, most of them have TAA or TAG as the stop codon, except three PCGs (COII, ND4, and Cytb) use an incomplete stop codon T. The phylogenetic tree based on the neighbour-joining method was constructed to provide relationship within Engraulidae, which could be a useful basis for management of this species.
The Hardenberg’s anchovy (Stolephorus insularis) is a species of the family Engraulidae which could be found from the Gulf of Aden east to Taiwan, south to western Indonesia (Fricke et al. Citation2012). Stolephorus insularis is fairly common throughout its range with high reproduction and used as bait in the tuna fishery in many areas (Whitehead et al. Citation1988). For the sake of marine biodiversity study and commercial value, to gain its molecular information, we determined and described the complete mitochondrial genome of S. insularis, and explored the phylogenetic relationship within Percoidea, which could be an effective genetic marker for such purposes.
The sample was collected from the coast nearby Taiwan Strait in Zhangzhou city, China (24°15′49″N; 118°06′31″ E) and stored in the laboratory of Zhejiang Ocean University with accession number 20180725PZ25. Codoncode Aligner was used for DNA sequences assembling. The calculation of base composition and phylogenetic construction was performed by MEGA6.0 software (Tamura et al. Citation2013).
The complete mitogenome of S. insularis is 16711 bp long (GeneBank Accession No. MH732976) consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and 2 main non-coding regions (Zhu et al. Citation2018). The overall base composition is 28.0% A, 26.8% C, 27.8% T and 17.4% G. There were 12 PCGs, 14 tRNA genes, and two rRNA genes located on the heavy strand, while one PCG (ND6) and eight tRNA genes (tRNA Gln, RNA Ala, tRNA Asn, tRNA Cys, tRNA Tyr, tRNA Ser, tRNA Glu, and tRNA Pro) on the light strand. Thirteen PCGs encode 3624 amino acids in total. All the protein-coding genes use the initiation codon ATG except COI uses GTG, which is quite common in vertebrate mtDNA (Miya et al. Citation2001). Most of them have TAA or TAG as the stop codon, except three PCGs (COII, ND4, and Cytb) ended with a single T, these incomplete termination codons were presumably completed as TAA by post-transcriptional polyadenylation (Ojala et al. Citation1981). There found totally 52 base pairs in 11 intergenic spacers with length from 1 to 10, and 47 base pairs in 12 overlapping areas.
The 12S rRNA and 16S rRNA are 952 and 1691 bp, which are both located in the common positions between tRNA-Phe and tRNA-Leu (UUA), separated by tRNA-Val (Wang and Xiuying Citation2008). Like any other vertebrate, the origin of OL is located in a cluster of five tRNA genes (WANCY) and The CR is located between the tRNA-Pro and tRNA-Phe genes with 1068 bp (Gong et al. Citation2017).
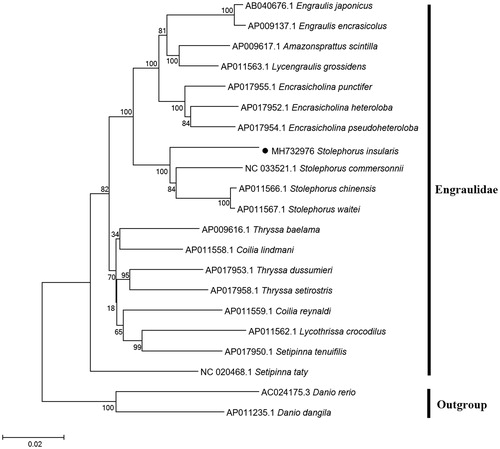
To explore the phylogenetic position of S. insularis, the NJ tree was constructed based on 12 mitochondrial PCGs located on a heavy strand of 19 species from Engraulidae family (). The phylogenetic tree indicated two major groups of this genus and showed that S. insularis has the closest relationship with S. commersonnii which meet the results based on morphology and molecular methods (Inoue et al. Citation2010).
Figure 1. Neighbour-joining (NJ) tree of 19 Engraulidae species based on 12 PCGs. The bootstrap values are based on 10,000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with a black spot.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Fricke R, Golani D, Appelbaum-Golani B. 2012. First record of the Indian Ocean anchovy Stolephorus insularis Hardenberg, 1933 (Clupeiformes: Engraulidae) in the mediterranean. Bioinvasions Rec. 1:303–306.
- Gong L, Liu L-Q, Guo B-Y, Ye Y-Y, Lü Z-M. 2017. The complete mitochondrial genome characterization of Thunnus obesus (Scombriformes: Scombridae) and phylogenetic analyses of Thunnus. Conservation Genet Resour. 9:379–383.
- Inoue JG, Miya M, Miller MJ, Sado T, Hanel R, Hatooka K, Aoyama J, Minegishi Y, Nishida M, Tsukamoto K. 2010. Deep-ocean origin of the freshwater eels. Biol Lett. 6:363–366.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondrial. Nature. 290:470.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang D, Xiuying W. 2008. A study on hydrological and hydraulic features of Trachidermus fasciatus migration. Adv Water Res Hydraulic Eng. 2:562–567.
- Whitehead P, Nelson G, Wongratana T. 1988. FAO species catalogue. Vol. 7. Clupeoid fishes of the world (Suborder Clupeoidei). An annotated and illustrated catalogue of the herrings, sardines, pilchards, sprats, shads, anchovies and wolf-herrings. FAO Fish. 125:305–579.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.
