Abstract
The slug moth, Narosa nigrisigna Wileman (Lepidoptera: Limacodidae), is a major tea pest in China. We have sequenced the complete mitochondrial genome of N. nigrisigna. The entire genome is 15,292 bp in length (GenBank accession No. MH675969) with consisting of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a 377 bp A + T-rich region. The gene order is consistent with other sequenced mt genome of moths and butterflies in Ditrysia. The A + T-rich region contains a motif ‘ATAGA’ and a 16 bp poly-T stretch. Phylogenetic analysis was performed using 13 protein-coding genes with 17 moths showed that N. nigrisigna is more closely related to other slug moths in family Limacodidae.
The slug moth, Narosa nigrisigna Wileman, belonging to the family Limacodidae in the superfamily Zygaenoidea, occurs in China. It consumes tea leaves and causes considerable economic losses. In this study, larvae of N. nigrisigna were collected from a tea plantation in Nanchang, Jiangxi province, China in July 2016 and identified different species by morphology. Voucher specimens (#CQNKY-LE-03-01-01) were deposited at the Insect Collection, Tea Research Institute of Chongqing Academy of Agricultural Science, Chongqing, China.
The complete mitochondrial genome of N. nigrisigna is shown to be a typical closed-circular and double-stranded DNA molecule in size of 15,292 bp (GenBank accession MH675969). It is low level among all reported Lepidoptera species and the smallest mt genome among all sequenced Limacodidae species: Monema flavescens (15,396 bp) (Liu et al. Citation2016), Cnidocampa flavescens (15,406 bp) (Peng et al. Citation2017) and Parasa consocia (15,296 bp) (Liu et al. Citation2017). The overall nucleotide composition of the major strand of the slug moth mt genome as follows: A = 38.97% (5,960), C = 11.12% (1,700), G = 7.67% (1,173), and T = 42.24% (6,459), with a total A + T content of 81.21%, that is heavily biased toward A and T nucleotides. AT- and GC-skew of the whole J-strand of N. nigrisigna is –0.040 and –0.183, respectively.
The mt genome encodes all 37 genes usually found in animal mt genomes, including 13 protein-coding genes (PCG), two ribosomal RNAs and 22 transfer RNAs. The gene arrangement in the mitochondrial genome of N. nigrisigna is conserved as other butterflies and moths mt genome in Ditrysia. In the mt genome of N. nigrisigna, a total of 21 bp overlaps have been found at seven gene junctions of the genome (atp8 and atp6 share 7 nucleotides; atp6 and cox3 share a nucleotide; trnR and trnN share 3 nucleotides; trnS1 and trnE share 1 nucleotides; nad4 and nad4L share 4 nucleotides; trnI and trnQ share 3 nucleotides; and nad2 and trnW share 2 nucleotides). The mt genome is loose and has a total of 151 bp intergenic sequence without the putative A + T-rich region. The intergenic sequences are at 15 locations ranging from 1 to 50 bp, the longest one locates between trnQ and nad2. The A + T-rich region of N. nigrisigna mt genome is 377 bp long and located between the rrnS and trnM genes. The A + T content of this region is 96.82%, the highest level of each region in this mt genome. This region includes the motif ‘ATAGA’ and a 16 bp poly-T stretch.
All 22 tRNA genes usually found in the mt genomes of insects are present in N. nigrisigna, 14 tRNA genes are encoded by J-strand and the others encoded by the N-strand. The nucleotide length of tRNA genes is ranging from 63 bp (trnR) to 71 bp (trnK and trnA), and A + T content is ranging from 70.42% (trnK) to 92.75% (trnE). 21 tRNA genes have the conventional cloverleaf shaped secondary structure and trnS1 gene lacks the dihydrouridine (DHU) arm. The two rRNA genes have been identified on the N-strand in the N. nigrisigna mt genome: the rrnL gene locates between trnL1 and trnV genes, and the rrnS gene between the trnV gene and the A + T-rich region. The length of rrnL and rrnS genes was 1,357 bp and 777 bp, and their A + T content was 85.56% and 86.49%, respectively. The total length of all 13 protein-coding genes is 11,172 bp, which is accounting for 73.06% of the whole genome sequence. The A + T content of the 13 genes ranges from 71.98% (cox1) to 88.89% (atp8). Twelve of the 13 PCGs start with ATN codons (ATG for atp6, cox2, cox3, cob, nad1, and nad4L; ATT for atp8, and nad2-3; ATA for nad4-6, and cox1 used CGA as start codon which has the same situation exists in most Lepidoptera species (Chen et al. Citation2015). Three PCGs (cox1-2 and nad4) have incomplete terminal codons consisting of single T nucleotide, and the other PCGs stop with TAA. The incomplete stop codon T is common and usually be demonstrated by the tRNA punctuation model, also could generate functional stop codons in polycistronic transcriptional division and polyadenylation mechanisms could (Ojala et al. Citation1981; Boore Citation2001; Stewart and Beckenbach Citation2009).
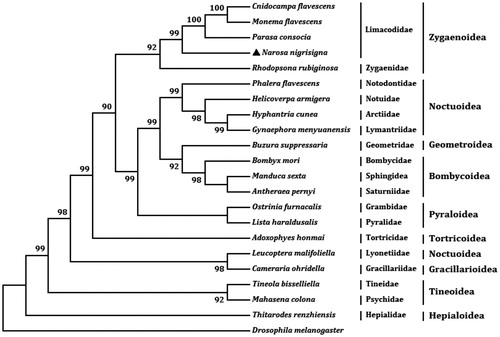
We analyzed nucleotides sequences of 13 PCGs with maximum likelihood (ML) method to understand the phylogenetic relationship of N. nigrisigna with other moths. The mt genome sequence of Drosophila melanogaster (GenBank accession no. DMU37541) was used as an outgroup. N. nigrisigna, C. flavescens, M. flavescens, and P. consocia belong to the family Limacodidae and they are clustered into a branch with 99% bootstrap value. Meanwhile, the two families Limacodidae and Zygaenidae (Rhodopsona rubiginosa) belong to the superfamily Zygaenoidea and they are clustered into a same branch in the phylogenetic tree ().
Figure 1. The maximum likelihood (ML) phylogenetic tree of Narosa nigrisigna and other moths. The GenBank accession numbers used for tree constructed are as follows: Cnidocampa flavescens (KY628213), Monema flavescens (KU946971), Parasa consocia (KX108765), Rhodopsona rubiginosa (KM244668), Phalera flavescens (JF440342), Helicoverpa armigera (GU188273), Hyphantria cunea (GU592049), Gynaephora menyuanensis (KC185412), Buzura suppressaria (KP278206), Bombyx mori (AF149768), Manduca sexta (EU286785), Antheraea pernyi (HQ264055), Ostrinia furnacalis (AF467260), Lista haraldusalis (KF709449), Adoxophyes honmai (DQ073916), Leucoptera malifoliella (JN790955), Cameraria ohridella (KJ508042), Tineola bisselliella (KJ508045), Mahasena colona (KY856825) and Thitarodes renzhiensis (HM744694).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18:1413–1416.
- Chen SC, Wang XQ, Wang JJ, Hu X, Peng P. 2015. The complete mitochondrial genome of a tea pest looper, Buzura suppressaria (Lepidoptera: Geometridae). Mitochondrial DNA Part A. 27:1–3154.
- Liu QN, Xin ZZ, Bian DD, Chai XY, Zhou CL, Tang BP. 2016. The first complete mitochondrial genome for the subfamily Limacodidae and implications for the higher phylogeny of Lepidoptera. Scientific Rep. 6:35878.
- Liu QN, Xin ZZ, Zhu XY, Chai XY, Zhao XM, Zhou CL, Tang BP. 2017. A transfer RNA gene rearrangement in the lepidopteran mitochondrial genome. Biochem Biophys Res Communs. 489:149–154.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Peng SY, Zhang Y, Zhang XC, Li Y, Huang ZR, Zhang YF, Zhang X, Ding JH, Geng XX, Li J. 2017. Complete mitochondrial genome of Cnidocampa flavescens (Lepidoptera: Limacodidae). Mitochondrial DNA Part B. 2:534–535.
- Stewart JB, Beckenbach AT. 2009. Characterization of mature mitochondrial transcripts in Drosophila, and the implications for the tRNA punctuation model in arthropods. Gene. 445:49–57.
