Abstract
Senna occidentalis (family: Fabaceae) is an annual herbaceous plant native to the Americans. The complete mitochondrial genome sequence of S. occidentalis is 447,106 bp long and contains 53 annotated genes, including 33 protein-coding genes, 17 tRNA genes, and 3 rRNA genes. Phylogenetic analysis of the mitochondrial genome confirmed divergence of S. occidentalis (subfamily: Caesalpinioideae) from the related subfamily Faboideae.
The genus Senna of the Fabaceae family includes about 260 species in South America (Marazzi et al. Citation2006). Senna occidentalis (formerly Cassia occidentalis) tissues are used in many countries as anti-inflammatory, antiplatelet, muscle relaxant, antimalaria, and antihemolytic medicines (Neboh and Ufelle Citation2015). A sample for sequencing was obtained from the National Agrobiodiversity Center (voucher number: IT 175099; geographic coordinates: N 35° 49′ 19″, E 127° 8′ 56″) in Jeonju, Korea.
Whole genome sequencing was performed using the Illumina Genome Analyzer platform (HiSeq4000, Illumina, USA). Reads were assembled using CLC Genomics Workbench, version 4.6 (CLC Inc., Aarhus, Denmark). The resulting three contigs were joined using the mitochondrial sequence of Cicer arientinum (GenBank: EU835853) as a reference. The draft genome was confirmed by paired-end read mapping, and gene annotation was conducted using DOGMA (Wyman et al. Citation2004) and BLAST (https://blast.ncbi.nlm.nih.gov/).
The mitochondrial genome of S. occidentalis consists of a single circular chromosome 447,106 bp in length. The genome contains 53 annotated genes, including 33 protein-coding genes, 17 tRNA genes, and 3 rRNA genes. Among these, we identified 11 open reading frames corresponding to the nad1 and nad5 genes. Interestingly, the nad1 and nad5 splice sites are located near exons 1, 2, 3, 4, and 5. The complete mitochondrial genome of S. occidentalis was deposited into GenBank (accession number MF358694).
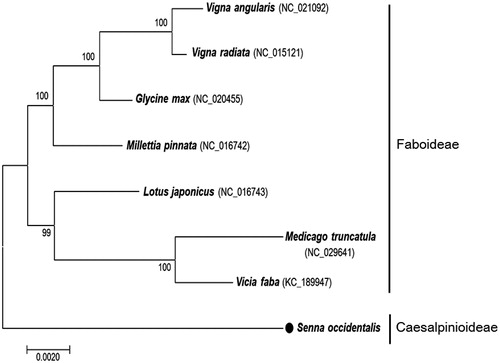
A phylogenetic tree of species in the Fabaceae family, including S. occidentalis, was constructed from mitochondrial sequences. Phylogenetic relationships were inferred from 16 common mitochondrial protein-coding genes. Genome alignments were performed using CLC Genomics Workbench, version 6.5 (CLC Inc., Aarhus, Denmark), and MEGA 7.0 (Kumar et al. Citation2016) was used for tree construction. Of the surveyed species, S. occidentalis was found to be most closely related to the last common ancestor of the Fabaceae family (). Phylogenetic analysis also confirmed placement of S. occidentalis in the subfamily Caesalpinioideae, as it is clearly divergent from members of the Faboideae subfamily.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Marazzi B, Endress PK, Queiroz LP, Conti E. 2006. Phylogenetic relationships within Senna (Leguminosae, Cassiinae) based on three chloroplast DNA regions: patterns in the evolution of floral symmetry and extrafloral nectaries. Am J Bot. 93:288–303.
- Neboh EE, Ufelle SA. 2015. Myeloprotective activity of crude methanolic leaf extract of Cassia occidentalis in cyclophosphamide-induced bone marrow suppression in Wistar rats. Adv Biomed Res. 4:5–8.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.