Abstract
Syzygium forrestii Merr. et Perry is an evergreen broad-leaved tree, 8–15 metres tall. It is mainly distributed in the Southwest of China (Nu river valley to Xishuangbanna area), growing from 800 to 2400 metres above sea level. It grows in the evergreen broad-leaved forest and the hillside shrubbery. It is an endemic plant species in China. At present, it has not been artificially introduced and cultivated. However, its complete plastome information and systematic position is still unknown. Here we report and characterize the complete plastid genome sequence of S. forrestii in an effort to provide genomic resources useful for promoting its conservation and systematic research. The complete plastome is 159,996 bp in length and contains the typical structure and gene content of angiosperm plastome, including two inverted repeat (IR) regions of 26,414 bp, a large single-copy (LSC) region of 88,560 bp and a small single-copy (SSC) region of 18,608 bp. The plastome contains 115 genes, consisting of 81 unique protein-coding genes, 30 unique tRNA gene and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA and 16S rRNA). The overall A/T content in the plastome of S. forrestii is 63.10%. The complete plastome sequence of S. forrestii will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Myrtaceae.
Introduction
Syzygium forrestii is an evergreen broad-leaved tree, 8–15 metres tall. It is mainly distributed in southwestern China (the Nu river valley to Xishuangbanna area), growing from 800 to 2,400 metres above sea level. It grows in the evergreen broad-leaved forest and the hillside shrubbery. It is an endemic plant species in China. At present, it has not been artificially introduced and cultivated, its complete plastome information and systematic position is still unknown. Here we report and characterize the complete plastid genome sequence of S. forrestii in an effort to provide genomic resources for promoting its conservation and systematic research. S.forrestii was sampled from the Ruili City in Yunnan Province of China (100.25°E, 26.86°N). A voucher specimen (specimen code: T. Yang et al., RL0700) was deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (HUTB), Hainan University, Haikou, China.
Around 6 Gb clean data were assembled against the plastome of Acca sellowiana (KX289887.1) (Machado et al. Citation2017) using MITO bim v1.8 (Hahn et al. Citation2013). The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of S. cumini (GQ870669.3). The annotation was corrected with DOGMA (Wyman et al. Citation2004).
The plastome of S. forrestii is found to possess a total length 159,996 bp with the typical quadripartite structure of angiosperms, contains two inverted repeats (IRs) of 26,414 bp, a large single-copy (LSC) region of 88,560 bp and a small single-copy (SSC) region of 18,608 bp. The plastome contains 115 genes, consisting of 81 unique protein-coding genes (seven of which are duplicated in the IR: rpl2, rpl23, ycf2, ycf15, ndhB, rps7 and rps12), 30 unique tRNA genes (seven of which are duplicated in the IR) and 4 unique rRNA genes (5S rRNA, 4.5S rRNA, 23S rRNA and 16S rRNA). Among these genes, 3 pseudogenes [Infa (translation from 84,286 to 84,212 bp), ycf15 (translation from 97,733 bp to 98,276 bp and translation from 150,824 to 150,281 bp), and ycf1 (translation from 113,878 bp to 114,981 bp)], 15 genes possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as atypical of angiosperms. The overall A/T content in the plastome of S. forrestii is 63.10%, which the corresponding value of the LSC, SSC and IR region were 65.50%, 69.20% and 57.40%, respectively.
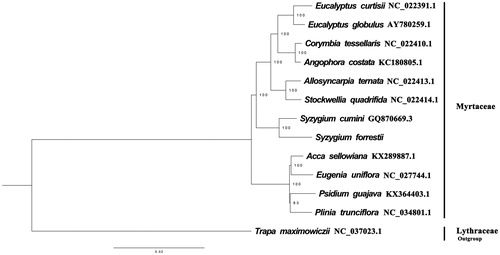
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of three published complete plastomes of Myrtaceae. Trapa maximowiczii (Lythraceae, Myrtales) as an outgroup. The phylogenetic analysis indicates that S. forrestii is more closer to S. cumini than other species in this study (). Most nodes in the plastome ML tree were strongly supported. The complete plastome sequence of S. forrestii will provide a useful resource for the conservation genetics of this species as well as for the phylogenetic studies of Myrtaceae.
Figure 1. The best ML phylogeny recovered from 14 complete plastome sequences by RAxML. Accession numbers: Syzygium forrestii (This study, Genbank accession number: MK102721), Acca sellowiana KX289887.1, Syzygium cumini NC_037023.1, Trapa maximowiczii NC_037023.1, Plinia trunciflora NC_034801.1,Psidium guajava KX364403.1, Eugenia uniflora NC_027744.1, Acca sellowiana KX289887.1, Stockwellia quadrifida NC_022414.1, Allosyncarpia ternata NC_022413.1, Angophora costata KC180805.1, Corymbia tessellaris NC_022410.1, Eucalyptus globulus AY780259.1, Eucalyptus curtisii NC_022391.1.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Machado LO, Vieira LD, Stefenon VM, Oliveira Pedrosa F, Souza EM, Guerra MP, Nodari RO. 2017. Phylogenomic relationship of feijoa (Acca sellowiana (O. Berg) Burret) with other Myrtaceae based on complete chloroplast genome sequences. Genetica. 145(2):163–174.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
