Abstract
The cutlassfish, Trichiurus haumela (Scombriformes: Trichiuridae), is well adapted to benthopelagic life with high commercial value in the world. In this study, mitogenome of this species was assembled with high coverage using Illumina sequencing data and the 16,855 bp length was acquired. The base composition is 28.75% for A, 29.33% for C, 16.14% for G, and 25.78% for T. The mitogenome includes 13 protein-coding genes, 21 transfer RNA, and two ribosomal RNA genes. The phylogeny showed that it is closely related to Trichiurus japonicus with high bootstrap value supported. The complete mitogenome of T. haumela can provide essential DNA molecular data for further phylogenetic and evolutionary analysis.
Trichiuridae in suborder Scombroidei of Perciformes, comprising three subfamilies, 10 genus, and 39 species, features very elongate and strongly compressed body with a small forked or hair-like caudal fin and is well adapted to benthopelagic life with high commercial value in the world (Nakamura and Parin Citation1993; Nelson Citation2006).
Samples were collected from Ningde, East China Sea (26.39°N, 119.31°E). Specimens were preserved in absolute alcohol and deposited in College of Life Science, Ningde Normal University, Ningde city, Fujian Province, China. Genomic DNA was extracted from muscle tissues following the manufacturer’s instruction in the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). Sequencing was conducted using the Illumina HiSeq platform (San Diego, CA). The sequence was preliminarily aligned within the CLUSTAL X program in BioEdit software (Thompson et al. Citation1997; Hall Citation1999). Protein-coding genes (PCGs), rRNA, genes were predicted by using MITOS tools (Bernt et al. Citation2013), and tRNA were done through tRNAscan-SE (Lowe and Chan Citation2016).
The complete mitogenome of Trichiurus haumela is 16,855 bp long in size (GenBank MH846121). The base composition is 28.75% for A, 29.33% for C, 16.14% for G, and 25.78% for T. Complete mitogenome of Trichiurus haumela contains 13 PCGs, two ribosomal RNA genes and a major non-coding region known as the CR (control region). Different with typical 22 tRNA invertebrate mitogenome, the mitogenome contained 21 transfer RNA genes except for tRNAPro. J-strand codes twelve PCGs (NAD1-5, NAD4L, COX1-3, CYTB, ATP6, and ATP8), two rRNAs (16S rRNA and 12S rRNA) and 14 tRNAs, while N-strand codes one PCGs (NAD6) and seven tRNAs. Gene arrangement of mitogenome for Trichiurus haumela is identical to the most common type of vertebrate (Liu and Cui Citation2009).
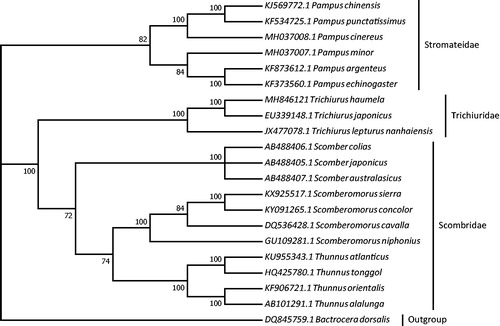
To validate the phylogenetic position, the mitogenomes of 19 species in order Scombriformes and an outgroup Bactrocera dorsalis (Diptera: Tephritidae) were clustered together with order to construct ML tree by using the maximum likelihood method based on the Tamura-Nei model in MEGA 7 software (Tamura and Nei Citation1993; Kumar et al. Citation2016). The tree inferred from 500 replicates was taken to represent the phylogeny of the species analyzed in this study (Felsenstein Citation1985). The result indicated that Trichiurus haumela was closely related to Trichiurus japonicus (Perciformes: Trichiuridae) with high bootstrap value supported (). Furthermore, three clades were correctly identified as assigned and monophyletic family Stromateidae, Trichiuridae, and Scombridae with high bootstrap confidence (). In conclusion, the mitochondrial genome of T. haumela deduced in present study can provide essential DNA molecular data for further phylogenetic and evolutionary analysis.
Figure 1. Molecular phylogeny of Trichiurus haumela and the related species in order Scombriformes based on complete mitogenome. Phylogenic tree is constructed by maximum likelihood method with 500 bootstrap replicates. Genbank accession number for tree construction is listed before the scientific name of species. The position of T. haumela is marked in solid square shape.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu Y, Cui Z. 2009. The complete mitochondrial genome sequence of the cutlassfish Trichiurus japonicus (Perciformes: Trichiuridae): genome characterization and phylogenetic considerations. Mar Genomics. 2:133–142.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucl Acids Res. 44:54–57.
- Nakamura I, Parin NV. 1993. Snake mackerels and cutlassfishes of the world (families Gempylidae and Trichiuridae). An annotated and illustrated catalogue of the snake mackerels, snoeks, escolars, gemfishes, sackfishes, domine, oilfish, cutlassfishes, scabbardfishes, hairtails and frostfishes known to date. FAO Species Catal. 125:1–136.
- Nelson JS. 2006. Fishes of the World. 4th ed. New York: John Wiley and Sons.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
