Abstract
Dolichos bean (Lablab purpureus) and Bambara groundnut (Vigna subterranea) are two traditional crops from Africa with important economic values. The study of such neglected or underused crops (orphan crops) should contribute to the resolution of food shortage in Africa. Here we assembled and described the complete plastid genome of the two Fabaceae orphan crop species. The length of the complete plastomes of L. purpureus and V. subterranea is 151,753 bp and 152,015 bp, respectively. Maximum-likelihood (ML) phylogenetic analyses indicated that L. purpureus and Phaseolus vulgaris are closely related, and V. subterranea and other Vigna species are clustered in one clade, which is congruent with former studies.
With the rapid growth of human population, food and nutrition shortage becomes a global challenge, and the utilization of unheeded or orphan crops potentially provides an alternative solution to this problem (Naluwairo Citation2011). Lablab purpureus (Dolichos bean) and Vigna subterranea (Bambara groundnut) are members of tribe Phaseoleae in Fabaceae, both of them are widely environmental adaptive and with the ability of nitrogen-fixing, regarded as important protein sources for African (Robotham and Chapman Citation2017; Gbaguidi et al. Citation2018). Here, we report the complete plastid genomes of the two neglected crops L. purpureus and V. subterranea on the basis of Illumina paired-end sequencing data.
In this study, fresh leaves were collected from plants that grow at the World AgroForestry Center (ICRAF) campus that are supported by the African Orphan Crops Consortium (AOCC). The total genomic DNA was extracted using a modified CTAB method (Sahu et al. Citation2012) and was used to construct the paired-end libraries with 250 bp insert size. Then the libraries were sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA), and 47 and 36 Gb high-quality reads were generated for L. purpureus, and V. subterranean, respectively. SOAPfilter v2.2 was used to trim the raw reads with the following criteria (1) reads with >10% base of N; (2) reads with >40% of low quality (quality value ≤10) sites; (3) reads contaminated by adaptor sequence or produced by PCR duplication. Finally, 40 Gb clean data of L. purpureus and 33 Gb clean data of V. subterranea were kept. Plastid genome was assembled based on NOVOPlasty v2.5.9 (Dierckxsens et al. Citation2017) using Arabidopsis thaliana plastid genome (NC_000932.1) as a seed. The plastome of Phaseolus vulgaris (DQ886273.1) and Vigna radiata (NC_013843.1) were used as references for the contig extending of L. purpureus and V. subterranea using the program MITO bim v1.8 (Hahn et al. Citation2013). The annotation of protein-coding genes was conducted with GeneWise v2.4.1 (Birney and Durbin Citation2000), and further verified on the web application GeSeq (Tillich et al. Citation2017). The annotation of transfer RNA (tRNA) and ribosomal RNA (rRNA) genes were performed using the program GeSeq (Tillich et al. Citation2017). The complete plastid genomes of L. purpureus and V. subterranea have been submitted to CNGB Nucleotide Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number: CNA0000811 and CNA0000812).
The complete plastid genome of L. purpureus and V. subterranea have a typical quadripartite structure, containing two inverted repeats (IRs), a large single-copy region (LSC), and a small single-copy region (SSC). L. purpureus has a total length of 151,753 bp, with two 26,054 bp IRs, a 81,727 bp LSC and a 17,918 bp SSC, while V. subterranea plastid genome is 152,015 bp long, with two 26,159 bp IRs, a 82,157 bp LSC, and a 17,540 bp SSC. For the gene content, both plastomes encode 71 protein-coding genes and four rRNA genes, L. purpureus has 32 tRNA genes, while V. subterranea has 33 tRNA genes. In both plastomes, twelve genes (rps19, rpl2, rpl23, ycf2, ndhB, rps7, rps12, trnN-GUU, trnR-ACG, trnA-UGC, trnV-GAC, trnL-CAA and trnM-CAU) are duplicated in the IR regions.
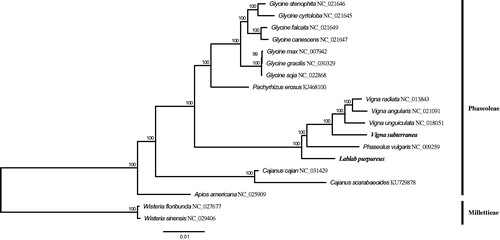
To reconstruct the phylogenetic relationships of the two orphan crops and other Fabaceae species, 17 published plastomes from tribes of Phaseoleae and Millettieae of Fabaceae were retrieved from the NCBI database. Seventy-seven protein-coding genes were extracted from all samples and used for gene alignment using the program MAFFT v7.017 (Katoh and Standley Citation2013). The aligned genes were concatenated into a data matrix of 67,020 sites. The Maximum-Likelihood tree was constructed using this concatenated dataset under the GTRCAT model using RAxML v8.2.4 (Stamatakis Citation2014) with 100 bootstraps. The phylogenetic analysis showed that L. purpureus and Phaseolus vulgaris were closely related, and V. subterranea and other Vigna species are clustered in the same clade (), which is congruent with former studies (Wang et al. Citation2017).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Birney E, Durbin R. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10:547–548.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Gbaguidi AA, Dansi A, Dossou-Aminon I, Gbemavo DSJC, Orobiyi A, Sanoussi F, Yedomonhan H. 2018. Agromorphological diversity of local Bambara groundnut (Vigna subterranea (L.) Verdc.) collected in Benin. Gen Res Crop Evol. 65:1159–1171.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Naluwairo R. 2011. Investing in Orphan Crops to Improve Food and Livelihood Security of Uganda’s Rural Poor: Policy Gaps, Opportunities and Recommendations. ACODE Policy Research Series. No. 43, 2011, Kampala, Uganda.
- Robotham O, Chapman M. 2017. Population genetic analysis of hyacinth bean (Lablab purpureus (L.) Sweet, Leguminosae) indicates an East African origin and variation in drought tolerance. Genetic Res Crop Evol. 64:139–148.
- Sahu SK, Thangaraj M, Kathiresan K. 2012. DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. ISRN Mol Biol. 2012:205049.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45:W6–W11.
- Wang YH, Qu XJ, Chen SY, Li DZ, Yi TS. 2017. Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Gen Genom. 13:41. doi:10.1007/s11295-017-1124-1