Abstract
To better understand the diversity and phylogeny of Scarabaeidae, we sequenced and annotated the nearly complete mitochondrial genome (mitogenome) of Amphimallon solstitiale (Coleoptera: Scarabaeidae: Melolonthinae). This mitogenome was 13,755 bp long and encoded 13 protein-coding genes (PCGs), 19 transfer RNA genes (tRNAs) and two ribosomal RNA unit genes. The A. solstitiale mitogenome with an A + T content of 69.45% presented a positive AT-skew (0.052) and a negative GC-skew (−0.236). Eleven PCGs started with a typical ATN codon, whereas the remaining two PCGs used CAC (cox1) and GTT (cox2) as the initial codon. All tRNAs had a typical secondary cloverleaf structure, except for trnS1 which lacked the dihydrouridine arm. The Bayesian phylogenetic analysis based on mitogenomic data recovered a phylogeny within Scarabaeidae as (Echirinae + (Scirtifomia + (Aphodiinae + (Melolonthinae + (Rutelinae + Cetoniinae))))).
Amphimallon solstitiale (Coleoptera: Scarabaeidae: Melolonthinae) is an underground pest which feeds on roots of potato, rape, and legume plants. Here, we sequenced and annotated the nearly complete mitochondrial genome (mitogenome) of A. solstitiale. Adult specimens were collected from Huining County, Gansu Province, China, in August 2017. Samples have been deposited in College of Pastoral Agricultural Science and Technology, Lanzhou University, Lanzhou, China. The total genomic DNA was extracted from a single specimen using a DNeasy Tissue Kit (Qiagen). The A. solstitiale mitogenome was amplified with a set of universal and specific primers, and sequenced in both directions.
We obtained the nearly complete mitogenome of A. solstitiale, with 13,755 bp long (GenBank accession number MH899179). The region that we failed to sequence in A. solstitiale was located between rrnS and nad2, and generally contained three transfer RNA genes (tRNAs; trnI, trnQ, and trnM) and a putative control region. This mitogenome encoded 13 protein-coding genes (PCGs), 19 tRNAs, two ribosomal RNA unit genes (rrnL and rrnS). All tRNAs had a typical secondary cloverleaf structure, except for trnS1 (AGN) which lacked the dihydrouridine arm. The order and orientation of the mitochondrial genes were identical to the inferred ancestral arrangement of insects (Boore Citation1999). Gene overlaps were found at seven gene junctions and involved a total of 27 bp, with the longest overlap (8 bp) between trnW and trnC, whereas a total of 51 bp intergenic spacers were presented in eight positions, ranging in size from 1 to 18 bp.
The A. solstitiale mitogenome with an A + T content of 69.45% showed a positive AT-skew (0.052) and a negative GC-skew (–0.236) on the J-strand. Among the 13 PCGs, the lowest A + T content was 63.32% in cox2, while the highest was 76.47% in atp8. Eleven PCGs started with a typical ATN codon: one (nad6) with ATC, two (nad3 and nad5) with ATA, three (nad2, atp8 and nad1) with ATT, five (atp6, cox3, nad4, nad4L, and cob) with ATG. The remaining two PCGs started with CAC (cox1) and GTT (cox2). Six PCGs terminated with TAA, and the remaining seven terminated with an incomplete stop codon TA or T.
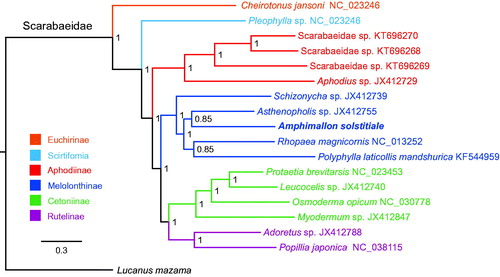
The concatenated nucleotide sequences of 13 PCGs from 17 Scarabaeidae species, representing six subfamilies, were used in phylogenetic analysis. Lucanus mazama from Lucanidae was used as an outgroup. We conducted a Bayesian phylogenetic analysis using the MrBayes 3.2.6 (Ronquist et al. Citation2012). The optimal partitioning schemes and corresponding nucleotide substitution models were determined by PartitionFinder 2.1.1 (Lanfear et al. Citation2017). Phylogenetic relationships among six subfamilies within Scarabaeidae were recovered as (Echirinae + (Scirtifomia + (Aphodiinae + (Melolonthinae + (Rutelinae + Cetoniinae))))) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.